[English] 日本語
 Yorodumi
Yorodumi- PDB-1o8a: Crystal Structure of Human Angiotensin Converting Enzyme (Native). -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1o8a | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal Structure of Human Angiotensin Converting Enzyme (Native). | |||||||||
 Components Components | ANGIOTENSIN CONVERTING ENZYME | |||||||||
 Keywords Keywords | METALLOPROTEASE / ACE / PEPTIDYL DIPEPTIDASE / TYPE-I MEMBRANE-ANCHORED PROTEIN. | |||||||||
| Function / homology |  Function and homology information Function and homology informationmononuclear cell proliferation / cell proliferation in bone marrow / bradykinin receptor binding / exopeptidase activity / regulation of angiotensin metabolic process / substance P catabolic process / tripeptidyl-peptidase activity / peptidyl-dipeptidase A / regulation of renal output by angiotensin / negative regulation of gap junction assembly ...mononuclear cell proliferation / cell proliferation in bone marrow / bradykinin receptor binding / exopeptidase activity / regulation of angiotensin metabolic process / substance P catabolic process / tripeptidyl-peptidase activity / peptidyl-dipeptidase A / regulation of renal output by angiotensin / negative regulation of gap junction assembly / metallodipeptidase activity / hormone catabolic process / bradykinin catabolic process / neutrophil mediated immunity / regulation of smooth muscle cell migration / regulation of hematopoietic stem cell proliferation / hormone metabolic process / mitogen-activated protein kinase kinase binding / mitogen-activated protein kinase binding / chloride ion binding / arachidonate secretion / post-transcriptional regulation of gene expression / peptide catabolic process / heart contraction / positive regulation of systemic arterial blood pressure / regulation of heart rate by cardiac conduction / regulation of systemic arterial blood pressure by renin-angiotensin / blood vessel remodeling / amyloid-beta metabolic process / hematopoietic stem cell differentiation / antigen processing and presentation of peptide antigen via MHC class I / peptidyl-dipeptidase activity / regulation of vasoconstriction / Metabolism of Angiotensinogen to Angiotensins / angiotensin maturation / metallocarboxypeptidase activity / blood vessel diameter maintenance / angiotensin-activated signaling pathway / kidney development / metalloendopeptidase activity / regulation of synaptic plasticity / regulation of blood pressure / male gonad development / metallopeptidase activity / peptidase activity / actin binding / endopeptidase activity / spermatogenesis / calmodulin binding / lysosome / endosome / negative regulation of gene expression / external side of plasma membrane / proteolysis / extracellular space / extracellular exosome / extracellular region / zinc ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å | |||||||||
 Authors Authors | Natesh, R. / Schwager, S.L.U. / Sturrock, E.D. / Acharya, K.R. | |||||||||
 Citation Citation |  Journal: Nature / Year: 2003 Journal: Nature / Year: 2003Title: Crystal structure of the human angiotensin-converting enzyme-lisinopril complex. Authors: Natesh, R. / Schwager, S.L. / Sturrock, E.D. / Acharya, K.R. | |||||||||
| History |
| |||||||||
| Remark 650 | HELIX DETERMINATION METHOD: AUTHOR PROVIDED. | |||||||||
| Remark 700 | SHEET DETERMINATION METHOD: AUTHOR PROVIDED. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1o8a.cif.gz 1o8a.cif.gz | 142 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1o8a.ent.gz pdb1o8a.ent.gz | 109.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1o8a.json.gz 1o8a.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/o8/1o8a https://data.pdbj.org/pub/pdb/validation_reports/o8/1o8a ftp://data.pdbj.org/pub/pdb/validation_reports/o8/1o8a ftp://data.pdbj.org/pub/pdb/validation_reports/o8/1o8a | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1o86SC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein / Sugars , 2 types, 7 molecules A

| #1: Protein | Mass: 68068.922 Da / Num. of mol.: 1 / Fragment: RESIDUES 68-656 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Organ: TESTIS / Production host: HOMO SAPIENS (human) / Organ: TESTIS / Production host:  References: UniProt: P22966, UniProt: P12821*PLUS, peptidyl-dipeptidase A |
|---|---|
| #2: Sugar | ChemComp-NAG / |
-Non-polymers , 5 types, 509 molecules 

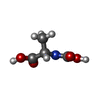






| #3: Chemical | ChemComp-ACT / | ||
|---|---|---|---|
| #4: Chemical | ChemComp-ZN / | ||
| #5: Chemical | ChemComp-NXA / | ||
| #6: Chemical | | #7: Water | ChemComp-HOH / | |
-Details
| Compound details | CONVERTS ANGIOTENSIN I TO ANGIOTENSIN II BY RELEASE OF THE TERMINAL HIS-LEU, THIS RESULTS IN AN ...CONVERTS ANGIOTENSI |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.4 Å3/Da / Density % sol: 49 % | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 4.7 / Details: PEG4000/SODIUM ACETATE., pH 4.70 | ||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 16 ℃ / Method: vapor diffusion | ||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SRS SRS  / Beamline: PX14.1 / Wavelength: 1.488 / Beamline: PX14.1 / Wavelength: 1.488 |
| Detector | Type: ADSC CCD / Detector: CCD / Date: Feb 25, 2002 / Details: MIRRORS |
| Radiation | Monochromator: SI / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.488 Å / Relative weight: 1 |
| Reflection | Resolution: 2→50 Å / Num. obs: 42782 / % possible obs: 98.8 % / Redundancy: 7.1 % / Biso Wilson estimate: 23.9 Å2 / Rmerge(I) obs: 0.081 / Net I/σ(I): 22.46 |
| Reflection shell | Resolution: 2→2.07 Å / Redundancy: 6.5 % / Rmerge(I) obs: 0.432 / Mean I/σ(I) obs: 4.44 / % possible all: 95.7 |
| Reflection | *PLUS Highest resolution: 2 Å |
| Reflection shell | *PLUS Highest resolution: 2 Å / % possible obs: 95.7 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1O86 Resolution: 2→47.14 Å / Cross valid method: THROUGHOUT / σ(F): 0
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 23.4 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→47.14 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor Rfree: 0.2208 / Rfactor Rwork: 0.1829 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller












 PDBj
PDBj







