| Entry | Database: PDB / ID: 1n1n
|
|---|
| Title | Structure of Mispairing of the Deoxycytosine with Deoxyadenosine 5' to the 8,9-Dihydro-8-(N7-guanyl)-9-Hydroxy-Aflatoxin B1 Adduct |
|---|
 Components Components | - 5'-D(*AP*CP*AP*TP*CP*GP*AP*TP*CP*T)-3'
- 5'-D(*AP*GP*AP*TP*CP*AP*AP*TP*GP*T)-3'
|
|---|
 Keywords Keywords | DNA / Aflatoxin B1 adduct 5' to AC mismatch / Major conformation |
|---|
| Function / homology | 8,9-DIHYDRO-9-HYDROXY-AFLATOXIN B1 / DNA Function and homology information Function and homology information |
|---|
| Method | SOLUTION NMR / distance geometry simulated annealing NMR data : Felix 97, 2000 molecular dynamics : XPLOR, AMBER matrix relaxation CORMA torsion angle dynamics Gaussian 98, INSIGHTII (2000) |
|---|
| Model type details | minimized average |
|---|
 Authors Authors | Stone, M.P. / Giri, I. |
|---|
 Citation Citation |  Journal: Biochemistry / Year: 2003 Journal: Biochemistry / Year: 2003
Title: Wobble dC.dA pairing 5' to the cationic guanine N7 8,9-dihydro-8-(N7-guanyl)-9-hydroxyaflatoxin B1 adduct: implications for nontargeted AFB1 mutagenesis.
Authors: Giri, I. / Stone, M.P. |
|---|
| History | | Deposition | Oct 18, 2002 | Deposition site: RCSB / Processing site: RCSB |
|---|
| Revision 1.0 | Oct 28, 2003 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Apr 28, 2008 | Group: Version format compliance |
|---|
| Revision 1.2 | Jul 13, 2011 | Group: Version format compliance |
|---|
| Revision 1.3 | Feb 23, 2022 | Group: Data collection / Database references / Derived calculations
Category: database_2 / pdbx_nmr_software ...database_2 / pdbx_nmr_software / pdbx_nmr_spectrometer / pdbx_struct_assembly / pdbx_struct_oper_list / struct_conn / struct_site
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_nmr_software.name / _pdbx_nmr_spectrometer.model / _struct_conn.pdbx_leaving_atom_flag / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id |
|---|
| Revision 1.4 | May 22, 2024 | Group: Data collection / Category: chem_comp_atom / chem_comp_bond |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Authors
Authors Citation
Citation Journal: Biochemistry / Year: 2003
Journal: Biochemistry / Year: 2003 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 1n1n.cif.gz
1n1n.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb1n1n.ent.gz
pdb1n1n.ent.gz PDB format
PDB format 1n1n.json.gz
1n1n.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads 1n1n_validation.pdf.gz
1n1n_validation.pdf.gz wwPDB validaton report
wwPDB validaton report 1n1n_full_validation.pdf.gz
1n1n_full_validation.pdf.gz 1n1n_validation.xml.gz
1n1n_validation.xml.gz 1n1n_validation.cif.gz
1n1n_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/n1/1n1n
https://data.pdbj.org/pub/pdb/validation_reports/n1/1n1n ftp://data.pdbj.org/pub/pdb/validation_reports/n1/1n1n
ftp://data.pdbj.org/pub/pdb/validation_reports/n1/1n1n Links
Links Assembly
Assembly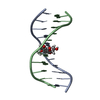
 Components
Components Sample preparation
Sample preparation Movie
Movie Controller
Controller










 PDBj
PDBj


