+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1k9w | ||||||
|---|---|---|---|---|---|---|---|
| Title | HIV-1(MAL) RNA Dimerization Initiation Site | ||||||
 Components Components | HIV-1 DIS(Mal)UU RNA | ||||||
 Keywords Keywords | RNA / HIV / hairpin | ||||||
| Function / homology | RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.1 Å MOLECULAR REPLACEMENT / Resolution: 3.1 Å | ||||||
 Authors Authors | Ennifar, E. / Walter, P. / Ehresmann, B. / Ehresmann, C. / Dumas, P. | ||||||
 Citation Citation |  Journal: Nat.Struct.Biol. / Year: 2001 Journal: Nat.Struct.Biol. / Year: 2001Title: Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Authors: Ennifar, E. / Walter, P. / Ehresmann, B. / Ehresmann, C. / Dumas, P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1k9w.cif.gz 1k9w.cif.gz | 57.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1k9w.ent.gz pdb1k9w.ent.gz | 43.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1k9w.json.gz 1k9w.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/k9/1k9w https://data.pdbj.org/pub/pdb/validation_reports/k9/1k9w ftp://data.pdbj.org/pub/pdb/validation_reports/k9/1k9w ftp://data.pdbj.org/pub/pdb/validation_reports/k9/1k9w | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 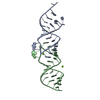
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: RNA chain | Mass: 7364.416 Da / Num. of mol.: 4 / Source method: obtained synthetically / Details: HIV-1(Mal) sequence with U265 and U287 mutations #2: Chemical | ChemComp-MG / |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.6 Å3/Da / Density % sol: 52.73 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 295 K / Method: vapor diffusion, hanging drop / pH: 7 Details: Isopropanol, magnesium chloride, spermine, cacodylate, pH 7.0, VAPOR DIFFUSION, HANGING DROP, temperature 295K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 20 ℃ / Method: vapor diffusion | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-3 / Wavelength: 0.93 Å / Beamline: ID14-3 / Wavelength: 0.93 Å |
| Detector | Type: MARRESEARCH / Detector: CCD |
| Radiation | Monochromator: Crystal / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.93 Å / Relative weight: 1 |
| Reflection | Resolution: 2.9→15 Å / Num. obs: 6955 / % possible obs: 96.3 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 2.9 % / Rsym value: 0.042 |
| Reflection shell | Resolution: 2.9→3.03 Å / Num. unique all: 909 / Rsym value: 0.296 / % possible all: 98.9 |
| Reflection | *PLUS Highest resolution: 3 Å / Lowest resolution: 20 Å / % possible obs: 95.9 % / Rmerge(I) obs: 0.037 |
| Reflection shell | *PLUS % possible obs: 99.4 % / Rmerge(I) obs: 0.104 / Mean I/σ(I) obs: 9.5 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 3.1→8 Å / Rfactor Rfree error: 0.013 / Data cutoff high absF: 422155.84 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 2 MOLECULAR REPLACEMENT / Resolution: 3.1→8 Å / Rfactor Rfree error: 0.013 / Data cutoff high absF: 422155.84 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 2
| |||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 10 Å2 / ksol: 0.09661481 e/Å3 | |||||||||||||||||||||||||
| Displacement parameters | Biso mean: 64.2 Å2
| |||||||||||||||||||||||||
| Refine analyze |
| |||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.1→8 Å
| |||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||
| LS refinement shell | Resolution: 3.1→3.28 Å / Rfactor Rfree error: 0.048 / Total num. of bins used: 6
| |||||||||||||||||||||||||
| Xplor file |
| |||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.227 / Rfactor Rfree: 0.265 / Rfactor Rwork: 0.227 | |||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||
| Displacement parameters | *PLUS | |||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| |||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor Rfree: 0.371 / Rfactor Rwork: 0.254 |
 Movie
Movie Controller
Controller











 PDBj
PDBj
































