+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1hvw | ||||||
|---|---|---|---|---|---|---|---|
| Title | HAIRPINLESS MUTANT OF OMEGA-ATRACOTOXIN-HV1A | ||||||
 Components Components | OMEGA-ATRACOTOXIN-HV1A | ||||||
 Keywords Keywords | TOXIN / cystine knot / beta-hairpin | ||||||
| Function / homology | Omega-atracotoxin / Omega-atracotoxin, conserved site / Omega-atracotoxin / Omega-atracotoxin (ACTX) type 1 family signature. / calcium channel inhibitor activity / defense response / toxin activity / extracellular region / Omega-hexatoxin-Hv1a Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / Torsion angle dynamics, dynamical simulated annealing | ||||||
 Authors Authors | Fletcher, J.I. / King, G.F. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2001 Journal: J.Biol.Chem. / Year: 2001Title: Functional significance of the beta hairpin in the insecticidal neurotoxin omega-atracotoxin-Hv1a. Authors: Tedford, H.W. / Fletcher, J.I. / King, G.F. #1:  Journal: Nat.Struct.Biol. / Year: 1997 Journal: Nat.Struct.Biol. / Year: 1997Title: The Structure of a Novel Insecticidal Neurotoxin, Omega-Atracotoxin-HV1, from the Venom of an Australian Funnel Web Spider Authors: Fletcher, J.I. / Smith, R. / O'Donoghue, S.I. / Nilges, M. / Connor, M. / Howden, M.E. / Christie, M.J. / King, G.F. #2:  Journal: Eur.J.Biochem. / Year: 1999 Journal: Eur.J.Biochem. / Year: 1999Title: Structure-function Studies of Omega-atracotoxin, A Potent Antagonist of Insect Voltage-gated Calcium Channels Authors: Wang, X. / Smith, R. / Fletcher, J.I. / Wilson, H. / Wood, C.J. / Howden, M.E. / King, G.F. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1hvw.cif.gz 1hvw.cif.gz | 115.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1hvw.ent.gz pdb1hvw.ent.gz | 78.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1hvw.json.gz 1hvw.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1hvw_validation.pdf.gz 1hvw_validation.pdf.gz | 333 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1hvw_full_validation.pdf.gz 1hvw_full_validation.pdf.gz | 413.9 KB | Display | |
| Data in XML |  1hvw_validation.xml.gz 1hvw_validation.xml.gz | 7.8 KB | Display | |
| Data in CIF |  1hvw_validation.cif.gz 1hvw_validation.cif.gz | 12.8 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hv/1hvw https://data.pdbj.org/pub/pdb/validation_reports/hv/1hvw ftp://data.pdbj.org/pub/pdb/validation_reports/hv/1hvw ftp://data.pdbj.org/pub/pdb/validation_reports/hv/1hvw | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 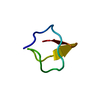
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 2623.899 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: This peptide was chemically synthesized. The native peptide is naturally found in Hadronyche versuta (Blue mountain funnel-web spider).The mutant hairpinless toxin was synthesized by solid- ...Details: This peptide was chemically synthesized. The native peptide is naturally found in Hadronyche versuta (Blue mountain funnel-web spider).The mutant hairpinless toxin was synthesized by solid-phase peptide synthesis, oxidized/folded in a glutathione redox buffer, then purified using reverse-phase HPLC. References: UniProt: P56207 |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||||||
| NMR details | Text: This structure was determined using standard 2D homonuclear techniques. |
- Sample preparation
Sample preparation
| Details |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions | Ionic strength: 0.005 / pH: 4.9 / Pressure: 1 atm / Temperature: 298 K | |||||||||
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Bruker DRX / Manufacturer: Bruker / Model: DRX / Field strength: 600 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: Torsion angle dynamics, dynamical simulated annealing Software ordinal: 1 Details: The structures are based on a total of 231 NOE-derived distance restraints, 19 dihedral-angle restraints, and 16 restraints defining 8 hydrogen bonds. | ||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 100 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller







 PDBj
PDBj X-PLOR
X-PLOR