| 登録情報 | データベース: PDB / ID: 1fuk
|
|---|
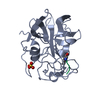
| タイトル | CRYSTAL STRUCTURE OF THE CARBOXY TERMINAL DOMAIN OF YEAST EIF4A |
|---|
 要素 要素 | EUKARYOTIC INITIATION FACTOR 4A |
|---|
 キーワード キーワード | TRANSLATION / Helicase / Initiation Factor 4A / DEAD-Box Protein |
|---|
| 機能・相同性 |  機能・相同性情報 機能・相同性情報
Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S / positive regulation of formation of translation preinitiation complex / Deadenylation of mRNA / eukaryotic translation initiation factor 4F complex / cytoplasmic translational initiation / ATP-dependent activity, acting on RNA / regulation of translational initiation / Translation initiation complex formation / Ribosomal scanning and start codon recognition / L13a-mediated translational silencing of Ceruloplasmin expression ...Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S / positive regulation of formation of translation preinitiation complex / Deadenylation of mRNA / eukaryotic translation initiation factor 4F complex / cytoplasmic translational initiation / ATP-dependent activity, acting on RNA / regulation of translational initiation / Translation initiation complex formation / Ribosomal scanning and start codon recognition / L13a-mediated translational silencing of Ceruloplasmin expression / translation initiation factor activity / translational initiation / cytoplasmic stress granule / RNA helicase activity / RNA helicase / ribosome / ATP hydrolysis activity / RNA binding / ATP binding / plasma membrane / cytoplasm類似検索 - 分子機能 ATP-dependent RNA helicase eIF4A, DEAD-box helicase domain / DEAD-box subfamily ATP-dependent helicases signature. / ATP-dependent RNA helicase DEAD-box, conserved site / RNA helicase, DEAD-box type, Q motif / DEAD-box RNA helicase Q motif profile. / DEAD/DEAH box helicase domain / DEAD/DEAH box helicase / Helicase conserved C-terminal domain / helicase superfamily c-terminal domain / Superfamilies 1 and 2 helicase C-terminal domain profile. ...ATP-dependent RNA helicase eIF4A, DEAD-box helicase domain / DEAD-box subfamily ATP-dependent helicases signature. / ATP-dependent RNA helicase DEAD-box, conserved site / RNA helicase, DEAD-box type, Q motif / DEAD-box RNA helicase Q motif profile. / DEAD/DEAH box helicase domain / DEAD/DEAH box helicase / Helicase conserved C-terminal domain / helicase superfamily c-terminal domain / Superfamilies 1 and 2 helicase C-terminal domain profile. / Superfamilies 1 and 2 helicase ATP-binding type-1 domain profile. / DEAD-like helicases superfamily / Helicase, C-terminal / Helicase superfamily 1/2, ATP-binding domain / P-loop containing nucleotide triphosphate hydrolases / Rossmann fold / P-loop containing nucleoside triphosphate hydrolase / 3-Layer(aba) Sandwich / Alpha Beta類似検索 - ドメイン・相同性 |
|---|
| 生物種 |   Saccharomyces cerevisiae (パン酵母) Saccharomyces cerevisiae (パン酵母) |
|---|
| 手法 |  X線回折 / X線回折 /  シンクロトロン / 解像度: 1.75 Å シンクロトロン / 解像度: 1.75 Å |
|---|
 データ登録者 データ登録者 | Caruthers, J.M. / Johnson, E.R. / McKay, D.B. |
|---|
 引用 引用 |  ジャーナル: Proc.Natl.Acad.Sci.USA / 年: 2000 ジャーナル: Proc.Natl.Acad.Sci.USA / 年: 2000
タイトル: Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase.
著者: Caruthers, J.M. / Johnson, E.R. / McKay, D.B. |
|---|
| 履歴 | | 登録 | 2000年9月15日 | 登録サイト: RCSB / 処理サイト: RCSB |
|---|
| 改定 1.0 | 2000年11月29日 | Provider: repository / タイプ: Initial release |
|---|
| 改定 1.1 | 2008年4月27日 | Group: Version format compliance |
|---|
| 改定 1.2 | 2011年7月13日 | Group: Version format compliance |
|---|
| 改定 1.3 | 2017年10月11日 | Group: Data collection / カテゴリ: reflns / Item: _reflns.percent_possible_obs |
|---|
| 改定 1.4 | 2024年2月7日 | Group: Data collection / Database references / Derived calculations
カテゴリ: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_struct_conn_angle / struct_conn / struct_site
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr1_symmetry / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.ptnr3_symmetry / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr1_symmetry / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_conn.ptnr2_symmetry / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id |
|---|
|
|---|
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報
 X線回折 /
X線回折 /  シンクロトロン / 解像度: 1.75 Å
シンクロトロン / 解像度: 1.75 Å  データ登録者
データ登録者 引用
引用 ジャーナル: Proc.Natl.Acad.Sci.USA / 年: 2000
ジャーナル: Proc.Natl.Acad.Sci.USA / 年: 2000 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 1fuk.cif.gz
1fuk.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb1fuk.ent.gz
pdb1fuk.ent.gz PDB形式
PDB形式 1fuk.json.gz
1fuk.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード 1fuk_validation.pdf.gz
1fuk_validation.pdf.gz wwPDB検証レポート
wwPDB検証レポート 1fuk_full_validation.pdf.gz
1fuk_full_validation.pdf.gz 1fuk_validation.xml.gz
1fuk_validation.xml.gz 1fuk_validation.cif.gz
1fuk_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/fu/1fuk
https://data.pdbj.org/pub/pdb/validation_reports/fu/1fuk ftp://data.pdbj.org/pub/pdb/validation_reports/fu/1fuk
ftp://data.pdbj.org/pub/pdb/validation_reports/fu/1fuk リンク
リンク 集合体
集合体
 要素
要素

 X線回折 / 使用した結晶の数: 2
X線回折 / 使用した結晶の数: 2  試料調製
試料調製 解析
解析 ムービー
ムービー コントローラー
コントローラー













 PDBj
PDBj





