[English] 日本語
 Yorodumi
Yorodumi- PDB-1fdn: REFINED CRYSTAL STRUCTURE OF THE 2[4FE-4S] FERREDOXIN FROM CLOSTR... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1fdn | ||||||
|---|---|---|---|---|---|---|---|
| Title | REFINED CRYSTAL STRUCTURE OF THE 2[4FE-4S] FERREDOXIN FROM CLOSTRIDIUM ACIDURICI AT 1.84 ANGSTROMS RESOLUTION | ||||||
 Components Components | FERREDOXIN | ||||||
 Keywords Keywords | ELECTRON TRANSPORT | ||||||
| Function / homology |  Function and homology information Function and homology information4 iron, 4 sulfur cluster binding / electron transfer activity / metal ion binding / cytoplasm Similarity search - Function | ||||||
| Biological species |  Clostridium acidurici (bacteria) Clostridium acidurici (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 1.84 Å X-RAY DIFFRACTION / Resolution: 1.84 Å | ||||||
 Authors Authors | Duee, E. / Fanchon, E. / Vicat, J. / Sieker, L.C. / Meyer, J. / Moulis, J-M. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1994 Journal: J.Mol.Biol. / Year: 1994Title: Refined crystal structure of the 2[4Fe-4S] ferredoxin from Clostridium acidurici at 1.84 A resolution. Authors: Duee, E.D. / Fanchon, E. / Vicat, J. / Sieker, L.C. / Meyer, J. / Moulis, J.M. #1:  Journal: Biochem.J. / Year: 1993 Journal: Biochem.J. / Year: 1993Title: Sequences of Clostridial Ferredoxins Authors: Meyer, J. / Moulis, J.-M. / Scherrer, N. / Gagnon, J. / Ulrich, J. #2:  Journal: J.Biol.Chem. / Year: 1988 Journal: J.Biol.Chem. / Year: 1988Title: Crystal Structure of Clostridium Acidi-Urici Ferredoxin at 5 Angstroms Resolution Based on Measurements of Anomalous X-Ray Scattering at Multiple Wavelengths Authors: Krishnamurthy, H.M. / Hendrickson, W.A. / Orme-Johnson, W.H. / Merritt, E.A. / Phizackerley, R.P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1fdn.cif.gz 1fdn.cif.gz | 22 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1fdn.ent.gz pdb1fdn.ent.gz | 14 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1fdn.json.gz 1fdn.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fd/1fdn https://data.pdbj.org/pub/pdb/validation_reports/fd/1fdn ftp://data.pdbj.org/pub/pdb/validation_reports/fd/1fdn ftp://data.pdbj.org/pub/pdb/validation_reports/fd/1fdn | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 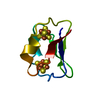
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Atom site foot note | 1: TWO ALTERNATE POSITIONS FOR THE SIDE CHAIN OF ASN 21 HAVE BEEN OBSERVED AND GIVEN OCCUPANCIES OF 0.5. 2: THE EXACT POSITIONS OF ASP 27 AND ASP 28 ARE UNDEFINED FROM THE X-RAY DATA. THESE RESIDUES HAVE BEEN BUILT WITH A GOOD GEOMETRY FOR THE MAIN CHAIN AND REASONABLE CHI ANGLES FOR THE SIDE CHAINS. |
- Components
Components
| #1: Protein | Mass: 5540.179 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Clostridium acidurici (bacteria) / References: UniProt: P00198 Clostridium acidurici (bacteria) / References: UniProt: P00198 | ||
|---|---|---|---|
| #2: Chemical | | #3: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2 Å3/Da / Density % sol: 38.49 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS Temperature: 281 K / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Radiation | Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| Reflection | *PLUS Highest resolution: 1.8 Å / Num. obs: 4203 / % possible obs: 92 % / Num. measured all: 15645 / Rmerge F obs: 0.062 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 1.84→18 Å / σ(F): 2 Details: THE EXACT POSITIONS OF ASP 27 AND ASP 28 ARE UNDEFINED FROM THE X-RAY DATA. THESE RESIDUES HAVE BEEN BUILT WITH A GOOD GEOMETRY FOR THE MAIN CHAIN AND REASONABLE CHI ANGLES FOR THE SIDE CHAINS.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.84→18 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Classification: refinement X-PLOR / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.169 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS Type: x_angle_d / Dev ideal: 2.58 |
 Movie
Movie Controller
Controller








 PDBj
PDBj










