[English] 日本語
 Yorodumi
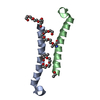
Yorodumi- PDB-1et1: CRYSTAL STRUCTURE OF HUMAN PARATHYROID HORMONE 1-34 AT 0.9 A RESO... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1et1 | ||||||
|---|---|---|---|---|---|---|---|
| Title | CRYSTAL STRUCTURE OF HUMAN PARATHYROID HORMONE 1-34 AT 0.9 A RESOLUTION | ||||||
 Components Components | PARATHYROID HORMONE | ||||||
 Keywords Keywords | HORMONE/GROWTH FACTOR / HELICAL DIMER / HORMONE-GROWTH FACTOR COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationmacromolecule biosynthetic process / parathyroid hormone receptor binding / type 1 parathyroid hormone receptor binding / negative regulation of bone mineralization involved in bone maturation / positive regulation of osteoclast proliferation / negative regulation of apoptotic process in bone marrow cell / response to parathyroid hormone / positive regulation of cell proliferation in bone marrow / hormone-mediated apoptotic signaling pathway / magnesium ion homeostasis ...macromolecule biosynthetic process / parathyroid hormone receptor binding / type 1 parathyroid hormone receptor binding / negative regulation of bone mineralization involved in bone maturation / positive regulation of osteoclast proliferation / negative regulation of apoptotic process in bone marrow cell / response to parathyroid hormone / positive regulation of cell proliferation in bone marrow / hormone-mediated apoptotic signaling pathway / magnesium ion homeostasis / positive regulation of signal transduction / response to fibroblast growth factor / cAMP metabolic process / phosphate ion homeostasis / Class B/2 (Secretin family receptors) / negative regulation of chondrocyte differentiation / response to vitamin D / peptide hormone receptor binding / positive regulation of inositol phosphate biosynthetic process / bone mineralization / positive regulation of glycogen biosynthetic process / bone resorption / response to cadmium ion / positive regulation of bone mineralization / Rho protein signal transduction / homeostasis of number of cells within a tissue / positive regulation of D-glucose import across plasma membrane / skeletal system development / hormone activity / response to lead ion / adenylate cyclase-activating G protein-coupled receptor signaling pathway / intracellular calcium ion homeostasis / cell-cell signaling / regulation of gene expression / response to ethanol / G alpha (s) signalling events / transcription by RNA polymerase II / receptor ligand activity / G protein-coupled receptor signaling pathway / response to xenobiotic stimulus / negative regulation of gene expression / positive regulation of gene expression / positive regulation of transcription by RNA polymerase II / extracellular space / extracellular region Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 0.9 Å SYNCHROTRON / Resolution: 0.9 Å | ||||||
 Authors Authors | Jin, L. / Briggs, S.L. / Chandrasekhar, S. / Chirgadze, N.Y. / Clawson, D.K. / Schevitz, R.W. / Smiley, D.L. / Tashjian, A.H. / Zhang, F. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2000 Journal: J.Biol.Chem. / Year: 2000Title: Crystal structure of human parathyroid hormone 1-34 at 0.9-A resolution. Authors: Jin, L. / Briggs, S.L. / Chandrasekhar, S. / Chirgadze, N.Y. / Clawson, D.K. / Schevitz, R.W. / Smiley, D.L. / Tashjian, A.H. / Zhang, F. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1et1.cif.gz 1et1.cif.gz | 47.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1et1.ent.gz pdb1et1.ent.gz | 35 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1et1.json.gz 1et1.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1et1_validation.pdf.gz 1et1_validation.pdf.gz | 360.5 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1et1_full_validation.pdf.gz 1et1_full_validation.pdf.gz | 360.7 KB | Display | |
| Data in XML |  1et1_validation.xml.gz 1et1_validation.xml.gz | 2.9 KB | Display | |
| Data in CIF |  1et1_validation.cif.gz 1et1_validation.cif.gz | 4.5 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/et/1et1 https://data.pdbj.org/pub/pdb/validation_reports/et/1et1 ftp://data.pdbj.org/pub/pdb/validation_reports/et/1et1 ftp://data.pdbj.org/pub/pdb/validation_reports/et/1et1 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Details | The biological functional unit is a monomer of hPTH (1-34). |
- Components
Components
| #1: Protein/peptide | Mass: 4125.778 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Production host: Homo sapiens (human) / Production host:  #2: Chemical | #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.76 Å3/Da / Density % sol: 30.08 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 4.5 Details: 2.5 M ammonium sulfate, 5% isopropanol and 0.1 M sodium acetate buffer, pH 4.5., VAPOR DIFFUSION, HANGING DROP, temperature 293K | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 20 ℃ | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 103 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 17-ID / Wavelength: 1 / Beamline: 17-ID / Wavelength: 1 |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Aug 10, 1998 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 0.9→13.5 Å / Num. all: 37771 / Num. obs: 37771 / % possible obs: 90.5 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 3 % / Biso Wilson estimate: 4.4 Å2 / Rmerge(I) obs: 0.054 / Net I/σ(I): 8.2 |
| Reflection shell | Resolution: 0.9→0.93 Å / Rmerge(I) obs: 0.163 / Num. unique all: 3148 / % possible all: 75.5 |
| Reflection | *PLUS Num. measured all: 111749 |
| Reflection shell | *PLUS % possible obs: 75.5 % |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 0.9→13.5 Å / σ(F): 0 / σ(I): 0 / Stereochemistry target values: Engh & Huber Details: Used conjugate-gradient algorithm with riding hydrogens with anisotropic B factor refinement.
| |||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 0.9→13.5 Å
| |||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||
| Software | *PLUS Name: SHELXL-97 / Classification: refinement | |||||||||||||||||||||||||
| Refinement | *PLUS σ(F): 0 / % reflection Rfree: 5 % / Rfactor Rfree: 0.14 | |||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||
| Displacement parameters | *PLUS | |||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller









 PDBj
PDBj


