[English] 日本語
 Yorodumi
Yorodumi- PDB-1egs: NMR STRUCTURE OF GROES MOBILE LOOP RESIDUES 19-27 IN THE SYNTHETI... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1egs | ||||||
|---|---|---|---|---|---|---|---|
| Title | NMR STRUCTURE OF GROES MOBILE LOOP RESIDUES 19-27 IN THE SYNTHETIC PEPTIDE (RESIDUES 13-32) BOUND TO GROEL, 20 STRUCTURES | ||||||
 Components Components | GROES | ||||||
 Keywords Keywords | CHAPERONIN / PROTEIN FOLDING / HEAT SHOCK | ||||||
| Function / homology |  Function and homology information Function and homology informationGroEL-GroES complex / virion assembly / protein folding chaperone / unfolded protein binding / protein-folding chaperone binding / protein folding / response to heat / ATP binding / metal ion binding / identical protein binding / cytosol Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method | SOLUTION NMR / MOLECULAR DYNAMICS, SIMULATED ANNEALING | ||||||
 Authors Authors | Landry, S.J. | ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 1996 Journal: Proc.Natl.Acad.Sci.USA / Year: 1996Title: Interplay of structure and disorder in cochaperonin mobile loops. Authors: Landry, S.J. / Taher, A. / Georgopoulos, C. / van der Vies, S.M. #1:  Journal: Nature / Year: 1993 Journal: Nature / Year: 1993Title: Characterization of a Functionally Important Mobile Domain of Groes Authors: Landry, S.J. / Zeilstra-Ryalls, J. / Fayet, O. / Georgopoulos, C. / Gierasch, L.M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1egs.cif.gz 1egs.cif.gz | 57.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1egs.ent.gz pdb1egs.ent.gz | 41.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1egs.json.gz 1egs.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/eg/1egs https://data.pdbj.org/pub/pdb/validation_reports/eg/1egs ftp://data.pdbj.org/pub/pdb/validation_reports/eg/1egs ftp://data.pdbj.org/pub/pdb/validation_reports/eg/1egs | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 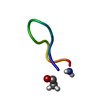
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 870.050 Da / Num. of mol.: 1 / Fragment: MOBILE LOOP Source method: isolated from a genetically manipulated source Source: (gene. exp.)  |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Sample conditions | pH: 6.5 / Temperature: 303 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Varian VXR500 / Manufacturer: Varian / Model: VXR500 / Field strength: 500 MHz |
|---|
- Processing
Processing
| NMR software |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: MOLECULAR DYNAMICS, SIMULATED ANNEALING / Software ordinal: 1 Details: REFINEMENT DETAILS CAN BE FOUND IN THE JRNL CITATION ABOVE. | |||||||||
| NMR ensemble | Conformers calculated total number: 20 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller







 PDBj
PDBj

