[English] 日本語
 Yorodumi
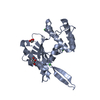
Yorodumi- PDB-1e0a: Cdc42 complexed with the GTPase binding domain of p21 activated kinase -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1e0a | ||||||
|---|---|---|---|---|---|---|---|
| Title | Cdc42 complexed with the GTPase binding domain of p21 activated kinase | ||||||
 Components Components |
| ||||||
 Keywords Keywords | SIGNALLING PROTEIN/KINASE / SIGNALLING PROTEIN / G PROTEIN SIGNALLING SER/THR KINASE / SIGNALLING PROTEIN-KINASE COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationCD28 dependent Vav1 pathway / VEGFR2 mediated vascular permeability / RHO GTPases activate PAKs / Sema3A PAK dependent Axon repulsion / CD209 (DC-SIGN) signaling / Generation of second messenger molecules / Ephrin signaling / RHOU GTPase cycle / RHOV GTPase cycle / RAC2 GTPase cycle ...CD28 dependent Vav1 pathway / VEGFR2 mediated vascular permeability / RHO GTPases activate PAKs / Sema3A PAK dependent Axon repulsion / CD209 (DC-SIGN) signaling / Generation of second messenger molecules / Ephrin signaling / RHOU GTPase cycle / RHOV GTPase cycle / RAC2 GTPase cycle / RHOH GTPase cycle / Smooth Muscle Contraction / MAPK6/MAPK4 signaling / RHOQ GTPase cycle / regulation of trans-synaptic signaling by endocannabinoid, modulating synaptic transmission / RAC1 GTPase cycle / observational learning / negative regulation of cell proliferation involved in contact inhibition / amygdala development / GBD domain binding / Golgi transport complex / positive regulation of pinocytosis / dendritic cell migration / endothelin receptor signaling pathway involved in heart process / gamma-aminobutyric acid secretion, neurotransmission / Regulation of actin dynamics for phagocytic cup formation / Signal transduction by L1 / cardiac neural crest cell migration involved in outflow tract morphogenesis / storage vacuole / glutamate secretion, neurotransmission / FCERI mediated MAPK activation / protein localization to cytoplasmic stress granule / apolipoprotein A-I receptor binding / positive regulation of epithelial cell proliferation involved in lung morphogenesis / neuron fate determination / regulation of attachment of spindle microtubules to kinetochore / organelle transport along microtubule / Inactivation of CDC42 and RAC1 / positive regulation of pseudopodium assembly / host-mediated perturbation of viral process / dendritic spine development / negative regulation of cell growth involved in cardiac muscle cell development / cardiac conduction system development / regulation of filopodium assembly / leading edge membrane / neuropilin signaling pathway / establishment of Golgi localization / EPHB-mediated forward signaling / positive regulation of microtubule nucleation / GTP-dependent protein binding / adherens junction organization / cell junction assembly / filopodium assembly / establishment of epithelial cell apical/basal polarity / dendritic spine morphogenesis / thioesterase binding / regulation of lamellipodium assembly / regulation of stress fiber assembly / gamma-tubulin binding / embryonic heart tube development / G beta:gamma signalling through CDC42 / hepatocyte growth factor receptor signaling pathway / positive regulation of vascular associated smooth muscle cell migration / RHO GTPases activate KTN1 / regulation of long-term synaptic potentiation / DCC mediated attractive signaling / regulation of postsynapse organization / CD28 dependent Vav1 pathway / positive regulation of fibroblast migration / Wnt signaling pathway, planar cell polarity pathway / positive regulation of filopodium assembly / RHOV GTPase cycle / phagocytosis, engulfment / regulation of mitotic nuclear division / nuclear migration / small GTPase-mediated signal transduction / Myogenesis / positive regulation of intracellular estrogen receptor signaling pathway / regulation of axonogenesis / branching morphogenesis of an epithelial tube / heart contraction / positive regulation of cytokinesis / spindle midzone / RHOJ GTPase cycle / neuromuscular junction development / RHOQ GTPase cycle / establishment of cell polarity / receptor clustering / Golgi organization / exocytosis / establishment or maintenance of cell polarity / dendrite development / RHOU GTPase cycle / transmission of nerve impulse / RHO GTPases activate PAKs / regulation of MAPK cascade / CDC42 GTPase cycle / macrophage differentiation / intercalated disc / RHOG GTPase cycle Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | SOLUTION NMR / DISTANCE GEOMETRY OF A SUBSTRUCTURE, CARTESIAN DYNAMICS | ||||||
 Authors Authors | Morreale, A. / Venkatesan, M. / Mott, H.R. / Owen, D. / Nietlispach, D. / Lowe, P.N. / Laue, E.D. | ||||||
 Citation Citation |  Journal: Nat.Struct.Biol. / Year: 2000 Journal: Nat.Struct.Biol. / Year: 2000Title: Solution Structure of Cdc42 Bound to the Gtpase Binding Domian of Pak Authors: Morreale, A. / Venkatesan, M. / Mott, H.R. / Owen, D. / Nietlispach, D. / Lowe, P.N. / Laue, E.D. | ||||||
| History |
| ||||||
| Remark 700 | SHEET DETERMINATION METHOD: THE SHEET STRUCTURE OF THIS MOLECULE IS BIFURCATED. IN ORDER TO ... SHEET DETERMINATION METHOD: THE SHEET STRUCTURE OF THIS MOLECULE IS BIFURCATED. IN ORDER TO REPRESENT THIS FEATURE IN THE SHEET RECORDS BELOW, TWO SHEETS ARE DEFINED. STRANDS 1, 2, 3 AND 4 OF SHEET A1 AND A2 ARE IDENTICAL. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1e0a.cif.gz 1e0a.cif.gz | 1.4 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1e0a.ent.gz pdb1e0a.ent.gz | 1.2 MB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1e0a.json.gz 1e0a.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/e0/1e0a https://data.pdbj.org/pub/pdb/validation_reports/e0/1e0a ftp://data.pdbj.org/pub/pdb/validation_reports/e0/1e0a ftp://data.pdbj.org/pub/pdb/validation_reports/e0/1e0a | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 20438.555 Da / Num. of mol.: 1 / Fragment: 1-184 / Mutation: YES Source method: isolated from a genetically manipulated source Details: COMPLEXED WITH 5'-GUANOSYL-IMIDO-TRIPHOSPHATE / Source: (gene. exp.)  Homo sapiens (human) / Cellular location: CYTOPLASM / Gene: CDC42 / Plasmid: PET16B / Cellular location (production host): CYTOPLASM / Gene (production host): CDC42 / Production host: Homo sapiens (human) / Cellular location: CYTOPLASM / Gene: CDC42 / Plasmid: PET16B / Cellular location (production host): CYTOPLASM / Gene (production host): CDC42 / Production host:  | ||
|---|---|---|---|
| #2: Protein/peptide | Mass: 5096.617 Da / Num. of mol.: 1 / Fragment: 75-118 / Mutation: YES Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: P35465, non-specific serine/threonine protein kinase | ||
| #3: Chemical | ChemComp-GNP / | ||
| #4: Chemical | ChemComp-MG / | ||
| #5: Water | ChemComp-HOH / | ||
| Compound details | CHAIN A: ENGINEERED| Sequence details | Q61L MUTANT. 7 C-TERMINAL RESIDUES REMOVED RESIDUES 73 AND 74 ARE FROM THE EXPRESSION | |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||
| NMR details | Text: SAMPLES WERE 13C,15N LABELLED CDC42 + UNLABELLED PAK OR 13C,15N LABELLED PAK + UNLABELLED CDC42 |
- Sample preparation
Sample preparation
| Details | Contents: 90% WATER, 10% D2O |
|---|---|
| Sample conditions | Ionic strength: 5 MM NA2HPO4, 25MM NACL / pH: 5.5 / Pressure: 1 atm / Temperature: 298 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Bruker DRX / Manufacturer: Bruker / Model: DRX / Field strength: 500 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: DISTANCE GEOMETRY OF A SUBSTRUCTURE, CARTESIAN DYNAMICS Software ordinal: 1 Details: REFINEMENT DETAILS CAN BE FOUND IN THE JRNL CITATION ABOVE | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: NO VIOLATIONS / Conformers calculated total number: 100 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller











 PDBj
PDBj






















