+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1dsk | ||||||
|---|---|---|---|---|---|---|---|
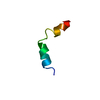
| Title | NMR SOLUTION STRUCTURE OF VPR59_86, 20 STRUCTURES | ||||||
 Components Components | VPR PROTEIN | ||||||
 Keywords Keywords | VIRAL PEPTIDE / POLYPEPTIDE | ||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated arrest of host cell cycle during G2/M transition / symbiont-mediated activation of host apoptosis / protein homooligomerization / virion component / viral penetration into host nucleus / host cell / monoatomic ion transmembrane transport / host extracellular space / DNA-templated transcription / regulation of DNA-templated transcription ...symbiont-mediated arrest of host cell cycle during G2/M transition / symbiont-mediated activation of host apoptosis / protein homooligomerization / virion component / viral penetration into host nucleus / host cell / monoatomic ion transmembrane transport / host extracellular space / DNA-templated transcription / regulation of DNA-templated transcription / symbiont entry into host cell / host cell nucleus Similarity search - Function | ||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | ||||||
| Method | SOLUTION NMR | ||||||
 Authors Authors | Yao, S. / Torres, A.M. / Azad, A.A. / Macreadie, I.G. / Norton, R.S. | ||||||
 Citation Citation |  Journal: J. Pept. Sci. / Year: 1998 Journal: J. Pept. Sci. / Year: 1998Title: Solution structure of peptides from HIV-1 Vpr protein that cause membrane permeabilization and growth arrest. Authors: Yao, S. / Torres, A.M. / Azad, A.A. / Macreadie, I.G. / Norton, R.S. #1: Journal: Gene / Year: 1996 Title: Characterization of a leucine-zipper-like domain in Vpr protein of human immunodeficiency virus type 1. Authors: Wang, L. / Mukherjee, S. / Narayan, O. / Zhao, L.J. #2: Journal: Proc. Natl. Acad. Sci. U.S.A. / Year: 1995 Title: A domain of human immunodeficiency virus type 1 Vpr containing repeated H(S/F)RIG amino acid motifs causes cell growth arrest and structural defects. Authors: Macreadie, I.G. / Castelli, L.A. / Hewish, D.R. / Kirkpatrick, A. / Ward, A.C. / Azad, A.A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1dsk.cif.gz 1dsk.cif.gz | 197.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1dsk.ent.gz pdb1dsk.ent.gz | 164 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1dsk.json.gz 1dsk.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ds/1dsk https://data.pdbj.org/pub/pdb/validation_reports/ds/1dsk ftp://data.pdbj.org/pub/pdb/validation_reports/ds/1dsk ftp://data.pdbj.org/pub/pdb/validation_reports/ds/1dsk | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 3353.068 Da / Num. of mol.: 1 / Fragment: RESIDUES 59 - 86 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Human immunodeficiency virus 1 / Genus: Lentivirus / References: UniProt: P12520 Human immunodeficiency virus 1 / Genus: Lentivirus / References: UniProt: P12520 |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|
- Sample preparation
Sample preparation
| Sample conditions | pH: 3.3 / Temperature: 298 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
- Processing
Processing
| Software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR software | Name:  X-PLOR / Version: 3.1 / Developer: BRUNGER / Classification: refinement X-PLOR / Version: 3.1 / Developer: BRUNGER / Classification: refinement | ||||||||||||
| NMR ensemble | Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller










 PDBj
PDBj