[English] 日本語
 Yorodumi
Yorodumi- PDB-1dsi: Solution structure of a duocarmycin sa-indole-alkylated dna dupleX -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1dsi | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solution structure of a duocarmycin sa-indole-alkylated dna dupleX | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA / DEOXYRIBONUCLEIC ACID / DUOCARMYCIN / MINOR GROOVE BINDING / ANTITUMOR AGENT / LIGAND-DNA COMPLEX | ||||||
| Function / homology | Chem-DSI / DNA / DNA (> 10) Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / NAB TO GENERATE A FAMILY OF CONFOMRERS THAT SAMPLE A WIDE RANGE OF CONFORMATIONAL SPACE. RESTRAINED MOLECULAR DYNAMICS TO GENERATE A RANGE OF STARTING STRUCTURES FOR DSI. PAIRS OF DNA, DSI STRUCTURES WERE THEN DOCKED, ITERATIVELY REFINED BY A SERIES OF RESTRAINED MOLECULAR DYNAMICS CALCULATIONS. | ||||||
 Authors Authors | Schnell, J.R. / Ketchem, R.R. / Boger, D.L. / Chazin, W.J. | ||||||
 Citation Citation | Journal: J.Am.Chem.Soc. / Year: 1999 Title: Binding-Induced Activation of DNA Alkylation by Duocarmycin SA: Insights from the Structure of an Indole Derivative-DNA Adduct Authors: Schnell, J.R. / Ketchem, R.R. / Boger, D.L. / Chazin, W.J. #1:  Journal: J.Mol.Biol. / Year: 1997 Journal: J.Mol.Biol. / Year: 1997Title: High Resolution Solution Structure of a DNA Duplex Alkylated by the Antitumor Agent Duocarmycin Sa Authors: Eis, P.S. / Smith, J.A. / Rydzewski, J.M. / Case, D.A. / Boger, D.L. / Chazin, W.J. #2:  Journal: J.Am.Chem.Soc. / Year: 1997 Journal: J.Am.Chem.Soc. / Year: 1997Title: Duocarmycin Sa Shortened, Simplified, and Extended Agents: A Systematic Examination of the Role of the DNA Binding Subunit Authors: Boger, D.L. / Herzog, D.L. / Bollinger, B. / Johnson, D.S. / Cai, H. / Goldberg, J. / Turnbull, P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1dsi.cif.gz 1dsi.cif.gz | 297.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1dsi.ent.gz pdb1dsi.ent.gz | 243.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1dsi.json.gz 1dsi.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ds/1dsi https://data.pdbj.org/pub/pdb/validation_reports/ds/1dsi ftp://data.pdbj.org/pub/pdb/validation_reports/ds/1dsi ftp://data.pdbj.org/pub/pdb/validation_reports/ds/1dsi | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 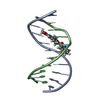
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 3357.223 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: MINOR GROOVE BOUND DUOCARMYCIN SA-INDOLE |
|---|---|
| #2: DNA chain | Mass: 3348.209 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: MINOR GROOVE BOUND DUOCARMYCIN SA-INDOLE |
| #3: Chemical | ChemComp-DSI / |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||||||||||
| NMR details | Text: THE STRUCTURE WAS DETERMINED USING HOMONUCLEAR 1H NMR DATA. RESONANCE ASSIGNMENTS WERE AIDED BY A 1H - 13C HSQC EXPERIMENT PERFORMED AT NATURAL ISOTOPE ABUNDANCE. |
- Sample preparation
Sample preparation
| Sample conditions | Ionic strength: 150mM / pH: 7 / Temperature: 300 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Bruker AMX600 / Manufacturer: Bruker / Model: AMX600 / Field strength: 600 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: NAB TO GENERATE A FAMILY OF CONFOMRERS THAT SAMPLE A WIDE RANGE OF CONFORMATIONAL SPACE. RESTRAINED MOLECULAR DYNAMICS TO GENERATE A RANGE OF STARTING STRUCTURES FOR DSI. PAIRS OF DNA, DSI ...Method: NAB TO GENERATE A FAMILY OF CONFOMRERS THAT SAMPLE A WIDE RANGE OF CONFORMATIONAL SPACE. RESTRAINED MOLECULAR DYNAMICS TO GENERATE A RANGE OF STARTING STRUCTURES FOR DSI. PAIRS OF DNA, DSI STRUCTURES WERE THEN DOCKED, ITERATIVELY REFINED BY A SERIES OF RESTRAINED MOLECULAR DYNAMICS CALCULATIONS. Software ordinal: 1 Details: REFINEMENT DETAILS CAN BE FOUND IN THE JRNL CITATION ABOVE. | ||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: LOWEST RESTRAINT VIOLATIONS / Conformers calculated total number: 40 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller







 PDBj
PDBj








































 HSQC
HSQC