[English] 日本語
 Yorodumi
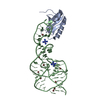
Yorodumi- PDB-1drz: U1A SPLICEOSOMAL PROTEIN/HEPATITIS DELTA VIRUS GENOMIC RIBOZYME C... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1drz | ||||||
|---|---|---|---|---|---|---|---|
| Title | U1A SPLICEOSOMAL PROTEIN/HEPATITIS DELTA VIRUS GENOMIC RIBOZYME COMPLEX | ||||||
 Components Components |
| ||||||
 Keywords Keywords | RNA BINDING PROTEIN/RNA / CATALYTIC RNA / RIBOZYME / RNA-BINDING PROTEIN / U1A / HDV / RNA BINDING PROTEIN-RNA COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationU1 snRNP binding / U1 snRNP / U1 snRNA binding / U4/U6 x U5 tri-snRNP complex / mRNA Splicing - Major Pathway / spliceosomal complex / mRNA splicing, via spliceosome / DNA binding / RNA binding / nucleoplasm ...U1 snRNP binding / U1 snRNP / U1 snRNA binding / U4/U6 x U5 tri-snRNP complex / mRNA Splicing - Major Pathway / spliceosomal complex / mRNA splicing, via spliceosome / DNA binding / RNA binding / nucleoplasm / identical protein binding / nucleus Similarity search - Function | ||||||
| Biological species |  Hepatitis delta virus Hepatitis delta virus Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MAD / Resolution: 2.3 Å MAD / Resolution: 2.3 Å | ||||||
 Authors Authors | Ferre-D'Amare, A.R. / Zhou, K. / Doudna, J.A. | ||||||
 Citation Citation |  Journal: Nature / Year: 1998 Journal: Nature / Year: 1998Title: Crystal structure of a hepatitis delta virus ribozyme. Authors: Ferre-D'Amare, A.R. / Zhou, K. / Doudna, J.A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1drz.cif.gz 1drz.cif.gz | 73.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1drz.ent.gz pdb1drz.ent.gz | 52.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1drz.json.gz 1drz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/dr/1drz https://data.pdbj.org/pub/pdb/validation_reports/dr/1drz ftp://data.pdbj.org/pub/pdb/validation_reports/dr/1drz ftp://data.pdbj.org/pub/pdb/validation_reports/dr/1drz | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||
| Unit cell |
|
- Components
Components
| #1: RNA chain | Mass: 23179.793 Da / Num. of mol.: 1 / Fragment: RIBOZYME DOMAIN Source method: isolated from a genetically manipulated source Details: RNA IS THE PRODUCT OF SELF-CLEAVAGE. NUCLEOTIDES 146 - 159 INCLUSIVE ARE AN ENGINEERED COGNATE BINDING SITE FOR THE U1A PROTEIN Source: (gene. exp.)  Hepatitis delta virus / Genus: Deltavirus Hepatitis delta virus / Genus: DeltavirusDescription: RNA PRODUCED BY IN VITRO RUN-OFF TRANSCRIPTION WITH BACTERIOPHAGE T7 RNA POLYMERASE FROM PLASMID DNA LINEARIZED WITH RESTRICTION ENZYME BSAI. T7 TRANSCRIPT Plasmid: PDU9 / Production host:  | ||||||
|---|---|---|---|---|---|---|---|
| #2: Protein | Mass: 11396.701 Da / Num. of mol.: 1 / Fragment: RNA BINDING DOMAIN / Mutation: Y31H, Q36R Source method: isolated from a genetically manipulated source Details: SELENOMETHIONYL PROTEIN / Source: (gene. exp.)  Homo sapiens (human) Homo sapiens (human)Description: (A1-98 Y31H Q36R) T7 PLASMID EXPRESSED IN E. COLI STRAIN B834 GROWN IN MINIMAL MEDIUM SUPPLEMENTED WITH SELENOMETHIONINE; Plasmid: T7 / Production host:  | ||||||
| #3: Chemical | | #4: Chemical | #5: Water | ChemComp-HOH / | Has protein modification | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 3 X-RAY DIFFRACTION / Number of used crystals: 3 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.17 Å3/Da / Density % sol: 67 % | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 7 / Details: pH 7.0 | ||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 25 ℃ / Method: vapor diffusion, sitting drop | ||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS NSLS  / Beamline: X4A / Wavelength: 0.9761, 0.9794, 0.9792, 1.1390 / Beamline: X4A / Wavelength: 0.9761, 0.9794, 0.9792, 1.1390 | |||||||||||||||
| Detector | Type: RIGAKU / Detector: IMAGE PLATE / Date: Mar 15, 1998 / Details: MIRROR | |||||||||||||||
| Radiation | Monochromator: SI / Protocol: MAD / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||
| Radiation wavelength |
| |||||||||||||||
| Reflection | Resolution: 2.9→41.3 Å / Num. all: 17817 / Num. obs: 17817 / % possible obs: 95.7 % / Observed criterion σ(I): 0 / Redundancy: 3.1 % / Biso Wilson estimate: 47.8 Å2 / Rmerge(I) obs: 0.065 / Net I/σ(I): 21.7 | |||||||||||||||
| Reflection shell | Resolution: 2.9→3 Å / Redundancy: 3.1 % / Rmerge(I) obs: 0.318 / Mean I/σ(I) obs: 5.2 / % possible all: 97.6 | |||||||||||||||
| Reflection | *PLUS Num. measured all: 55889 | |||||||||||||||
| Reflection shell | *PLUS % possible obs: 97.6 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MAD / Resolution: 2.3→20 Å / Rfactor Rfree error: 0.007 / Data cutoff high rms absF: 2082995.34 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 MAD / Resolution: 2.3→20 Å / Rfactor Rfree error: 0.007 / Data cutoff high rms absF: 2082995.34 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 43.3 Å2 / ksol: 0.336 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 76.2 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.3→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.3→2.44 Å / Rfactor Rfree error: 0.023 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: CNS / Version: 0.3 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 2.3 Å / Lowest resolution: 20 Å / σ(F): 0 / % reflection Rfree: 9.8 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS Biso mean: 76.2 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor Rfree: 0.399 / % reflection Rfree: 9.4 % / Rfactor Rwork: 0.426 |
 Movie
Movie Controller
Controller











 PDBj
PDBj


































