+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1c4e | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | GURMARIN FROM GYMNEMA SYLVESTRE | ||||||||||||
 Components Components | PROTEIN (GURMARIN) | ||||||||||||
 Keywords Keywords | PLANT PROTEIN / GURMARIN / SWEET TASTE SUPPRESSION / CYSTINE KNOT / SWEET TASTE TRANSDUCTION | ||||||||||||
| Function / homology | Gurmarin/antifungal peptide / Pro-gurmarin Function and homology information Function and homology information | ||||||||||||
| Biological species |  Gymnema sylvestre (plant) Gymnema sylvestre (plant) | ||||||||||||
| Method | SOLUTION NMR / DISTANCE GEOMETRY, DYNAMICAL SIMULATED ANNEALING | ||||||||||||
 Authors Authors | Fletcher, J.I. / Dingley, A.J. / King, G.F. | ||||||||||||
 Citation Citation |  Journal: Eur.J.Biochem. / Year: 1999 Journal: Eur.J.Biochem. / Year: 1999Title: High-resolution solution structure of gurmarin, a sweet-taste-suppressing plant polypeptide. Authors: Fletcher, J.I. / Dingley, A.J. / Smith, R. / Connor, M. / Christie, M.J. / King, G.F. #1:  Journal: J.Biomol.NMR / Year: 1995 Journal: J.Biomol.NMR / Year: 1995Title: Three-Dimensional Structure of Gurmarin, a Sweet Taste-Suppressing Polypeptide Authors: Arai, K. / Ishima, R. / Morikawa, S. / Miyasaka, A. / Imoto, T. / Yoshimura, S. / Aimoto, S. / Akasaka, K. #2:  Journal: J.Biochem.(Tokyo) / Year: 1992 Journal: J.Biochem.(Tokyo) / Year: 1992Title: Amino Acid Sequence of Sweet-Taste-Suppressing Peptide (Gurmarin) from the Leaves of Gymnema Sylvestre Authors: Kamei, K. / Takano, R. / Miyasaka, A. / Imoto, T. / Hara, S. #3:  Journal: COMP.BIOCHEM.PHYSIOL. A: PHYSIOL. / Year: 1991 Journal: COMP.BIOCHEM.PHYSIOL. A: PHYSIOL. / Year: 1991Title: A Novel Peptide Isolated from the Leaves of Gymnema Sylvestre-I. Characterization and its Suppressive Effect on the Neural Responses to Sweet Taste Stimuli in the Rat Authors: Imoto, T. / Miyasaka, A. / Ishima, R. / Akasaka, K. | ||||||||||||
| History |
|
- Structure visualization
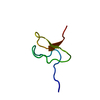
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1c4e.cif.gz 1c4e.cif.gz | 227.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1c4e.ent.gz pdb1c4e.ent.gz | 185.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1c4e.json.gz 1c4e.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/c4/1c4e https://data.pdbj.org/pub/pdb/validation_reports/c4/1c4e ftp://data.pdbj.org/pub/pdb/validation_reports/c4/1c4e ftp://data.pdbj.org/pub/pdb/validation_reports/c4/1c4e | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 4220.952 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Details: CHEMICALLY SYNTHESIZED / Source: (natural)  Gymnema sylvestre (plant) / Tissue: LEAVES / References: UniProt: P25810 Gymnema sylvestre (plant) / Tissue: LEAVES / References: UniProt: P25810 |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||
| NMR details | Text: THE STRUCTURE WAS DETERMINED USING STANDARD 2D HOMONUCLEAR TECHNIQUES |
- Sample preparation
Sample preparation
| Sample conditions | pH: 2.9 / Pressure: ATMOSPHERIC atm / Temperature: 298 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Bruker AMX600 / Manufacturer: Bruker / Model: AMX600 / Field strength: 600 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: DISTANCE GEOMETRY, DYNAMICAL SIMULATED ANNEALING / Software ordinal: 1 / Details: SEE PRIMARY REFERENCE ABOVE. | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: LOWEST ENERGY / Conformers calculated total number: 100 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller










 PDBj
PDBj
 ECOSY
ECOSY