+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1byj | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
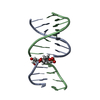
| Title | GENTAMICIN C1A A-SITE COMPLEX | |||||||||
 Components Components | RNA (16S RNA) | |||||||||
 Keywords Keywords | RNA / COMPLEX (AMINOGLYCOSIDE-RIBOSOMAL RNA) | |||||||||
| Function / homology | Chem-GE1 / 3,5-DIAMINO-CYCLOHEXANOL / Chem-GE3 / RNA / RNA (> 10) Function and homology information Function and homology information | |||||||||
| Method | SOLUTION NMR / simulated annealing | |||||||||
 Authors Authors | Yoshizawa, S. / Fourmy, D. / Puglisi, J.D. | |||||||||
 Citation Citation |  Journal: EMBO J. / Year: 1998 Journal: EMBO J. / Year: 1998Title: Structural origins of gentamicin antibiotic action. Authors: Yoshizawa, S. / Fourmy, D. / Puglisi, J.D. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1byj.cif.gz 1byj.cif.gz | 788.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1byj.ent.gz pdb1byj.ent.gz | 575.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1byj.json.gz 1byj.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/by/1byj https://data.pdbj.org/pub/pdb/validation_reports/by/1byj ftp://data.pdbj.org/pub/pdb/validation_reports/by/1byj ftp://data.pdbj.org/pub/pdb/validation_reports/by/1byj | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 8656.185 Da / Num. of mol.: 1 / Fragment: DECODING REGION / Source method: obtained synthetically / Details: 16S RNA WAS CHEMICALLY SYNTHESIZED. |
|---|---|
| #2: Sugar | ChemComp-GE1 / |
| #3: Chemical | ChemComp-GE2 / |
| #4: Sugar | ChemComp-GE3 / |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||
| NMR details | Text: A TOTAL OF 326 RNA, 7 GENTAMICIN AND 46 GENTAMICIN-RNA RESTRAINTS WERE USED. 111 EXPERIMENTAL DIHEDRAL RESTRAINTS WERE USED. FOR 38 THE FINAL STRUCTURES DEVIATIONS FROM EXPERIMENTAL RESTRAINTS ...Text: A TOTAL OF 326 RNA, 7 GENTAMICIN AND 46 GENTAMICIN-RNA RESTRAINTS WERE USED. 111 EXPERIMENTAL DIHEDRAL RESTRAINTS WERE USED. FOR 38 THE FINAL STRUCTURES DEVIATIONS FROM EXPERIMENTAL RESTRAINTS ARE: RMSD NMR DISTANCE RESTRAINTS (379) (A): 0.0249 (0.0002) RMSD NMR DIHEDRAL RESTRAINTS (111) (DEG): 0.0127 (0.0016) DEVIATIONS FROM DEALIZED GEOMETRY (CVFF FORCEFIELD) RMSD BONDS (A): 0.0260 ( 0.0000) RMSD ANGLES (DEG): 0.0621 (0.0001) RMSD IMPROPERS (DEG): 0.0485 ( 0.0025) HEAVY ATOM RMS DEVIATIONS OF AVERAGE, MINIMIZED STRUCTURE FROM BEST- FIT SUPERPOSITION OF THE 38 FINAL STRUCTURES RMSD ALL RNA + GENTAMICIN (A): 0.89 |
- Sample preparation
Sample preparation
| Sample conditions | Ionic strength: 10 mM SODIUM PHOSPHATE / pH: 6.4 / Temperature: 308 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 | ||||||||||||
| NMR ensemble | Conformer selection criteria: LEAST RESTRAINT VIOLATION / Conformers calculated total number: 38 / Conformers submitted total number: 38 |
 Movie
Movie Controller
Controller







 PDBj
PDBj