[English] 日本語
 Yorodumi
Yorodumi- PDB-1bn0: SL3 HAIRPIN FROM THE PACKAGING SIGNAL OF HIV-1, NMR, 11 STRUCTURES -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1bn0 | ||||||
|---|---|---|---|---|---|---|---|
| Title | SL3 HAIRPIN FROM THE PACKAGING SIGNAL OF HIV-1, NMR, 11 STRUCTURES | ||||||
 Components Components | SL3 RNA HAIRPIN | ||||||
 Keywords Keywords | RNA / HIV-1 / PACKAGING / TETRALOOP / RIBONUCLEIC ACID | ||||||
| Function / homology | RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / DISTANCE GEOMETRY, RESTRAINED MOLECULAR DYNAMICS | ||||||
 Authors Authors | Pappalardo, L. / Kerwood, D.J. / Pelczer, I. / Borer, P.N. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1998 Journal: J.Mol.Biol. / Year: 1998Title: Three-dimensional folding of an RNA hairpin required for packaging HIV-1. Authors: Pappalardo, L. / Kerwood, D.J. / Pelczer, I. / Borer, P.N. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1bn0.cif.gz 1bn0.cif.gz | 143.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1bn0.ent.gz pdb1bn0.ent.gz | 116.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1bn0.json.gz 1bn0.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bn/1bn0 https://data.pdbj.org/pub/pdb/validation_reports/bn/1bn0 ftp://data.pdbj.org/pub/pdb/validation_reports/bn/1bn0 ftp://data.pdbj.org/pub/pdb/validation_reports/bn/1bn0 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 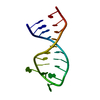
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 6477.919 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||
| NMR details | Text: THE STRUCTURES WERE DETERMINED FROM 2D NOESY EXPERIMENTS PERFORMED BOTH IN H2O AND D2O. THE DISTANCE RESTRAINTS DETERMINED FROM MARDIGRAS WERE BASED ON INTENSITIES FROM A NOESY WITH A MIXING ...Text: THE STRUCTURES WERE DETERMINED FROM 2D NOESY EXPERIMENTS PERFORMED BOTH IN H2O AND D2O. THE DISTANCE RESTRAINTS DETERMINED FROM MARDIGRAS WERE BASED ON INTENSITIES FROM A NOESY WITH A MIXING TIME OF 300MS. DQF-COSY AND H-P COSY WERE USED TO ASCERTAIN INFORMATION ABOUT THE TORSION ANGLES. |
- Sample preparation
Sample preparation
| Details | Contents: WATER |
|---|---|
| Sample conditions | Ionic strength: 30mM / pH: 7 / Pressure: NORMAL / Temperature: 303 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Bruker DRX500 / Manufacturer: Bruker / Model: DRX500 / Field strength: 500 MHz |
|---|
- Processing
Processing
| Software | Name:  AMBER / Classification: refinement AMBER / Classification: refinement | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR software |
| |||||||||||||||
| Refinement | Method: DISTANCE GEOMETRY, RESTRAINED MOLECULAR DYNAMICS / Software ordinal: 1 Details: AVERAGE SIXTH ROOT RESIDUAL NMR INDEX = 0.07, AVERAGE RESTRAINT VIOLATION = 0.06 ANGSTROM. | |||||||||||||||
| NMR ensemble | Conformer selection criteria: LEAST RESTRAINT VIOLATION / Conformers calculated total number: 500 / Conformers submitted total number: 11 |
 Movie
Movie Controller
Controller






 PDBj
PDBj






























