[English] 日本語
 Yorodumi
Yorodumi- PDB-1bds: DETERMINATION OF THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1bds | ||||||
|---|---|---|---|---|---|---|---|
| Title | DETERMINATION OF THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE ANTIHYPERTENSIVE AND ANTIVIRAL PROTEIN BDS-I FROM THE SEA ANEMONE ANEMONIA SULCATA. A STUDY USING NUCLEAR MAGNETIC RESONANCE AND HYBRID DISTANCE GEOMETRY-DYNAMICAL SIMULATED ANNEALING | ||||||
 Components Components | BDS-I | ||||||
 Keywords Keywords | ANTI-HYPERTENSIVE / ANTI-VIRAL PROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology informationnematocyst / ion channel inhibitor activity / sodium channel regulator activity / potassium channel regulator activity / regulation of blood pressure / toxin activity / extracellular region Similarity search - Function | ||||||
| Biological species |  Anemonia sulcata (snake-locks sea anemone) Anemonia sulcata (snake-locks sea anemone) | ||||||
| Method | SOLUTION NMR | ||||||
 Authors Authors | Clore, G.M. / Driscoll, P.C. / Gronenborn, A.M. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 1989 Journal: Biochemistry / Year: 1989Title: Determination of the three-dimensional solution structure of the antihypertensive and antiviral protein BDS-I from the sea anemone Anemonia sulcata: a study using nuclear magnetic resonance ...Title: Determination of the three-dimensional solution structure of the antihypertensive and antiviral protein BDS-I from the sea anemone Anemonia sulcata: a study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing. Authors: Driscoll, P.C. / Gronenborn, A.M. / Beress, L. / Clore, G.M. #1:  Journal: Biochemistry / Year: 1989 Journal: Biochemistry / Year: 1989Title: A Proton Nuclear Magnetic Resonance Study of the Antihypertensive and Antiviral Protein Bds-I from the Sea Anemone Anemonia Sulcata. Sequential and Stereospecific Resonance Assignment and Secondary Structure Authors: Driscoll, P.C. / Clore, G.M. / Beress, L. / Gronenborn, A.M. #2:  Journal: FEBS Lett. / Year: 1989 Journal: FEBS Lett. / Year: 1989Title: The Influence of Stereospecific Assignments on the Determination of Three-Dimensional Structures of Proteins by Nuclear Magnetic Resonance Spectroscopy. Application to the Sea Anemone Protein Bds-I Authors: Driscoll, P.C. / Gronenborn, A.M. / Clore, G.M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1bds.cif.gz 1bds.cif.gz | 25.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1bds.ent.gz pdb1bds.ent.gz | 16.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1bds.json.gz 1bds.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bd/1bds https://data.pdbj.org/pub/pdb/validation_reports/bd/1bds ftp://data.pdbj.org/pub/pdb/validation_reports/bd/1bds ftp://data.pdbj.org/pub/pdb/validation_reports/bd/1bds | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 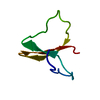
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Atom site foot note | 1: RESIDUES PRO 36 AND PRO 42 ARE CIS PROLINES. | |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 4719.434 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Anemonia sulcata (snake-locks sea anemone) Anemonia sulcata (snake-locks sea anemone)References: UniProt: P11494 |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|
- Processing
Processing
| Software |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| NMR software | Name:  XPLOR / Developer: BRUNGER / Classification: refinement XPLOR / Developer: BRUNGER / Classification: refinement | ||||||||
| Refinement | Software ordinal: 1 Details: REFINEMENT. THE METHOD USED TO DETERMINE AND REFINE THE STRUCTURE IS THE HYBRID METRIC MATRIX DISTANCE GEOMETRY-DYNAMICAL SIMULATED ANNEALING METHOD (M.NILGES, G.M.CLORE, A.M. GRONENBORN, ...Details: REFINEMENT. THE METHOD USED TO DETERMINE AND REFINE THE STRUCTURE IS THE HYBRID METRIC MATRIX DISTANCE GEOMETRY-DYNAMICAL SIMULATED ANNEALING METHOD (M.NILGES, G.M.CLORE, A.M. GRONENBORN, FEBS LETT. 229, 317-324 (1988)) USING THE PROGRAM XPLOR (A.T. BRUENGER, YALE UNIVERSITY, CT 06511). STRUCTURAL STATISTICS RMS DEVIATION FROM EXPERIMENTAL RESTRAINTS *(1)* RESTRAINT TYPE NUMBER OF RESTRAINTS RMS (ANGSTROMS) ALL 513 0.079 INTERRESIDUE SHORT RANGE 150 0.086 INTERRESIDUE LONG RANGE 105 0.100 INTRARESIDUE 234 0.065 HBOND *(2)* 24 0.043 POTENTIAL ENERGY TERMS TYPE ENERGY (KCAL/MOL) F(NOE) *(3)* 160 F(TOR) *(4)* 24 F(REPEL) *(5)* 72 LENNARD-JONES VAN DER WAALS ENERGY (E(L-J)) CALCULATED USING THE *CHARMM* EMPIRICAL ENERGY FUNCTION IS -117 KCAL/MOL. DEVIATIONS FROM IDEALIZED GEOMETRY *(6)* TYPE TOTAL NUMBER RMS DEVIATION BONDS 646 0.013 (ANGSTROMS) ANGLES 1157 2.517 (DEGREES) IMPROPERS 242 0.797 (DEGREES) NOTES. *(1)* THE RMS DEVIATION FROM THE EXPERIMENTAL RESTRAINTS ARE CALCULATED WITH RESPECT TO THE UPPER AND LOWER LIMITS OF THE DISTANCE RESTRAINTS. NONE OF THE STRUCTURES EXHIBITED VIOLATIONS GREATER THAN 0.5 ANGSTROMS. *(2)* FOR EACH BACKBONE HYDROGEN BOND THERE ARE TWO RESTRAINTS - R(NH-O) .LT. 2.3 ANGSTROMS AND R(N-O) .LT. 3.3 ANGSTROMS. THE LOWER LIMITS ARE GIVEN BY THE SUM OF THE VAN DER WAALS RADII OF THE RELEVANT ATOMS. *(3)* THE VALUES OF THE SQUARE-WELL NOE POTENTIAL F(NOE) ARE CALCULATED WITH A FORCE CONSTANT OF 50 KCAL/MOL/ANGSTROM**2. *(4)* THE VALUES OF F(PHI) ARE CALCULATED WITH A FORCE CONSTANT OF 200 KCAL/MOL/RAD**2. F(PHI) IS A SQUARE-WELL DIHEDRAL POTENTIAL WHICH IS USED TO RESTRICT THE RANGES OF 23 PHI AND 21 CHI1 TORSION ANGLES, AND THE OMEGA PEPTIDE BOND TORSION ANGLES OF THE FIVE PROLINE RESIDUES (PRO 36 AND 42 BEING RESTRAINED TO THE CIS CONFORMATION AND THE OTHERS TO TRANS). *(5)* THE VALUE OF THE VAN DER WAALS REPULSION TERM F(REPEL) IS CALCULATED WITH A FORCE CONSTANT OF 4 KCAL/MOL/ANGSTROM**4 WITH THE HARD SPHERE VAN DER WAALS RADII SET TO 0.8 TIMES THE STANDARD VALUES USED IN THE *CHARMM* EMPIRICAL ENERGY FUNCTION. *(6)* THE IMPROPER TERMS SERVE TO MAINTAIN PLANARITY AND APPROPRIATE CHIRALITY. THEY ALSO MAINTAIN THE PEPTIDE BONDS OF ALL RESIDUES (WITH THE EXCEPTION OF PROLINES) IN THE TRANS CONFORMATION. IN THE DYNAMICAL SIMULATED ANNEALING CALCULATIONS, THE RESTRAINTS FOR THE DISULFIDE BRIDGES ARE INCLUDED IN THE BOND AND ANGLE TERMS. A TOTAL OF 42 STRUCTURES CONSISTENT WITH THE NMR DATA WERE CALCULATED. THIS ENTRY REPRESENTS THE COORDINATES OBTAINED BY AVERAGING THE COORDINATES OF THE INDIVIDUAL STRUCTURES AND SUBJECTING THE RESULTING COORDINATES TO FURTHER RESTRAINED MINIMIZATION. THE COORDINATES OF THESE 42 STRUCTURES ARE GIVEN IN THE PDB ENTRY *2BDS*. THE THERMAL PARAMETERS GIVEN IN THIS ENTRY REPRESENT THE ATOMIC RMS DEVIATION OF THE INDIVIDUAL STRUCTURES ABOUT THE MEAN COORDINATE POSITIONS. ALL THE INTERPROTON DISTANCE AND TORSION ANGLE RESTRAINTS ARE AVAILABLE IN THE PDB NMR RESTRAINT FILE. | ||||||||
| NMR ensemble | Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller








 PDBj
PDBj