+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1b7d | ||||||
|---|---|---|---|---|---|---|---|
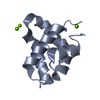
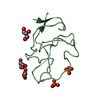
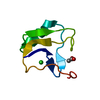
| Title | NEUROTOXIN (TS1) FROM BRAZILIAN SCORPION TITYUS SERRULATUS | ||||||
 Components Components | PROTEIN (NEUROTOXIN TS1) | ||||||
 Keywords Keywords | TOXIN / LONG-CHAIN NEUROTOXIN | ||||||
| Function / homology |  Function and homology information Function and homology informationsodium channel inhibitor activity / defense response to fungus / toxin activity / killing of cells of another organism / extracellular region Similarity search - Function | ||||||
| Biological species |  Tityus serrulatus (Brazilian scorpion) Tityus serrulatus (Brazilian scorpion) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.73 Å MOLECULAR REPLACEMENT / Resolution: 1.73 Å | ||||||
 Authors Authors | Polikarpov, I. / Sanches Jr., M.S. / Marangoni, S. / Toyama, M.H. / Teplyakov, A. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1999 Journal: J.Mol.Biol. / Year: 1999Title: Crystal structure of neurotoxin Ts1 from Tityus serrulatus provides insights into the specificity and toxicity of scorpion toxins. Authors: Polikarpov, I. / Junior, M.S. / Marangoni, S. / Toyama, M.H. / Teplyakov, A. #1:  Journal: Acta Crystallogr.,Sect.D / Year: 1998 Journal: Acta Crystallogr.,Sect.D / Year: 1998Title: Crystallisation and Preliminary Diffraction Data of Neurotoxin Ts-Gamma from the Venom of Scorpion Tityus Serrulatus Authors: Golubev, A.M. / Lee, W.H. / Marangoni, S. / Novello, J.C. / Oliveira, B. / Toyama, M.H. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1b7d.cif.gz 1b7d.cif.gz | 26.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1b7d.ent.gz pdb1b7d.ent.gz | 15.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1b7d.json.gz 1b7d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/b7/1b7d https://data.pdbj.org/pub/pdb/validation_reports/b7/1b7d ftp://data.pdbj.org/pub/pdb/validation_reports/b7/1b7d ftp://data.pdbj.org/pub/pdb/validation_reports/b7/1b7d | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1ahoS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 6901.012 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Tityus serrulatus (Brazilian scorpion) / Secretion: VENOM / References: UniProt: P15226 Tityus serrulatus (Brazilian scorpion) / Secretion: VENOM / References: UniProt: P15226 |
|---|---|
| #2: Chemical | ChemComp-PO4 / |
| #3: Water | ChemComp-HOH / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.81 Å3/Da / Density % sol: 32.84 % Description: DIFFRACTION DATA HAVE BEEN COLLECTED AT THE LABORATORIO NACIONAL DE LUZ SINCROTRON (LNLS), CAMPINAS, BRAZIL | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 6 / Details: pH 6.0 | ||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging dropDetails: 0.01ml protein solution was mixed with 0.005ml reservoir PH range low: 6.5 / PH range high: 5.2 | ||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 295 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  LNLS LNLS  / Beamline: D03B-MX1 / Wavelength: 1.37 / Beamline: D03B-MX1 / Wavelength: 1.37 |
| Detector | Type: MAR scanner 345 mm plate / Detector: IMAGE PLATE / Details: CYLINDRICAL MIRROR |
| Radiation | Monochromator: SI BENT SINGLE-CRYSTAL / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.37 Å / Relative weight: 1 |
| Reflection | Resolution: 1.73→13 Å / Num. obs: 5149 / % possible obs: 97.1 % / Redundancy: 2.6 % / Rsym value: 0.066 |
| Reflection shell | Resolution: 1.73→1.77 Å / Redundancy: 2.3 % / Mean I/σ(I) obs: 3 / Rsym value: 0.26 / % possible all: 81.6 |
| Reflection | *PLUS Num. measured all: 13402 / Rmerge(I) obs: 0.066 |
| Reflection shell | *PLUS % possible obs: 81.6 % / Rmerge(I) obs: 0.26 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1AHO Resolution: 1.73→13 Å / SU ML: 0.15 / Cross valid method: THROUGHOUT / σ(F): 0
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 21.1 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.73→13 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: REFMAC / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller










 PDBj
PDBj




