[English] 日本語
 Yorodumi
Yorodumi- PDB-1avn: HUMAN CARBONIC ANHYDRASE II COMPLEXED WITH THE HISTAMINE ACTIVATOR -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1avn | ||||||
|---|---|---|---|---|---|---|---|
| Title | HUMAN CARBONIC ANHYDRASE II COMPLEXED WITH THE HISTAMINE ACTIVATOR | ||||||
 Components Components | CARBONIC ANHYDRASE II | ||||||
 Keywords Keywords | LYASE / OXO-ACID | ||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of cellular pH reduction / positive regulation of dipeptide transmembrane transport / regulation of monoatomic anion transport / secretion / cyanamide hydratase / cyanamide hydratase activity / arylesterase activity / regulation of chloride transport / Reversible hydration of carbon dioxide / morphogenesis of an epithelium ...positive regulation of cellular pH reduction / positive regulation of dipeptide transmembrane transport / regulation of monoatomic anion transport / secretion / cyanamide hydratase / cyanamide hydratase activity / arylesterase activity / regulation of chloride transport / Reversible hydration of carbon dioxide / morphogenesis of an epithelium / angiotensin-activated signaling pathway / positive regulation of synaptic transmission, GABAergic / regulation of intracellular pH / carbonic anhydrase / carbonate dehydratase activity / carbon dioxide transport / Erythrocytes take up oxygen and release carbon dioxide / Erythrocytes take up carbon dioxide and release oxygen / neuron cellular homeostasis / apical part of cell / myelin sheath / extracellular exosome / zinc ion binding / plasma membrane / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  FOURIER SYNTHESIS / Resolution: 2 Å FOURIER SYNTHESIS / Resolution: 2 Å | ||||||
 Authors Authors | Briganti, F. / Mangani, S. / Orioli, P. / Scozzafava, A. / Vernaglione, G. / Supuran, C.T. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 1997 Journal: Biochemistry / Year: 1997Title: Carbonic anhydrase activators: X-ray crystallographic and spectroscopic investigations for the interaction of isozymes I and II with histamine. Authors: Briganti, F. / Mangani, S. / Orioli, P. / Scozzafava, A. / Vernaglione, G. / Supuran, C.T. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1avn.cif.gz 1avn.cif.gz | 70.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1avn.ent.gz pdb1avn.ent.gz | 49.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1avn.json.gz 1avn.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/av/1avn https://data.pdbj.org/pub/pdb/validation_reports/av/1avn ftp://data.pdbj.org/pub/pdb/validation_reports/av/1avn ftp://data.pdbj.org/pub/pdb/validation_reports/av/1avn | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 29157.863 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: COMPLEX WITH HISTAMINE / Source: (gene. exp.)  Homo sapiens (human) / Cell: ERYTHROCYTES / Cell line: BL21 / Plasmid: PACA/HCA II / Species (production host): Escherichia coli / Production host: Homo sapiens (human) / Cell: ERYTHROCYTES / Cell line: BL21 / Plasmid: PACA/HCA II / Species (production host): Escherichia coli / Production host:  |
|---|
-Non-polymers , 5 types, 159 molecules 







| #2: Chemical | ChemComp-ZN / |
|---|---|
| #3: Chemical | ChemComp-HG / |
| #4: Chemical | ChemComp-AZI / |
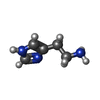
| #5: Chemical | ChemComp-HSM / |
| #6: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.15 Å3/Da / Density % sol: 42.68 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 7.7 Details: HCA II CRYSTALLIZED FROM 2.3 M AMMONIUM SULFATE, 50MM TRIS-HCL, 3MM SODIUM AZIDE, PH 7.7 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 4 ℃ / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 293 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: MACSCIENCE M18X / Wavelength: 1.5418 ROTATING ANODE / Type: MACSCIENCE M18X / Wavelength: 1.5418 |
| Detector | Type: SIEMENS / Detector: AREA DETECTOR / Date: Nov 1, 1996 |
| Radiation | Monochromator: GRAPHITE(002) / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2→30.9 Å / Num. obs: 14777 / % possible obs: 81 % / Redundancy: 3.4 % / Rmerge(I) obs: 0.076 / Rsym value: 0.09 / Net I/σ(I): 33.61 |
| Reflection shell | Resolution: 2→2.25 Å / Redundancy: 2.3 % / Rmerge(I) obs: 0.064 / Mean I/σ(I) obs: 10.7 / Rsym value: 0.174 / % possible all: 73 |
| Reflection | *PLUS Num. measured all: 49849 / Rmerge(I) obs: 0.089 |
| Reflection shell | *PLUS % possible obs: 73 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  FOURIER SYNTHESIS / Resolution: 2→15 Å FOURIER SYNTHESIS / Resolution: 2→15 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 14.9 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.15 Å / Luzzati d res low obs: 30 Å / Luzzati sigma a obs: 0.15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→15 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  CCP4 / Classification: refinement CCP4 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.154 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller






















 PDBj
PDBj


















