+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9874 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of PaaZ with NADPH | |||||||||||||||
 Map data Map data | This map is of PaaZ with NADPH bound. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | substrate channeling / bi-functional enzyme / hydrolase / dehydrogenase | |||||||||||||||
| Function / homology |  Function and homology information Function and homology information3-oxo-5,6-dehydrosuberyl-CoA semialdehyde dehydrogenase / oxepin-CoA hydrolase / ether hydrolase activity / hydrolase activity, acting on carbon-carbon bonds, in ketonic substances / oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor / phenylacetate catabolic process / oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor / enoyl-CoA hydratase activity / identical protein binding Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
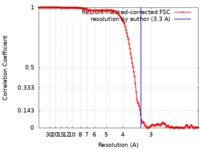
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||||||||
 Authors Authors | Gakher L / Cannone G / Katagihallimath N / Sowdhamini R / Sathyanarayanan N | |||||||||||||||
| Funding support |  India, India,  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway. Authors: Nitish Sathyanarayanan / Giuseppe Cannone / Lokesh Gakhar / Nainesh Katagihallimath / Ramanathan Sowdhamini / Subramanian Ramaswamy / Kutti R Vinothkumar /    Abstract: Substrate channeling is a mechanism for the internal transfer of hydrophobic, unstable or toxic intermediates from the active site of one enzyme to another. Such transfer has previously been ...Substrate channeling is a mechanism for the internal transfer of hydrophobic, unstable or toxic intermediates from the active site of one enzyme to another. Such transfer has previously been described to be mediated by a hydrophobic tunnel, the use of electrostatic highways or pivoting and by conformational changes. The enzyme PaaZ is used by many bacteria to degrade environmental pollutants. PaaZ is a bifunctional enzyme that catalyzes the ring opening of oxepin-CoA and converts it to 3-oxo-5,6-dehydrosuberyl-CoA. Here we report the structures of PaaZ determined by electron cryomicroscopy with and without bound ligands. The structures reveal that three domain-swapped dimers of the enzyme form a trilobed structure. A combination of small-angle X-ray scattering (SAXS), computational studies, mutagenesis and microbial growth experiments suggests that the key intermediate is transferred from one active site to the other by a mechanism of electrostatic pivoting of the CoA moiety, mediated by a set of conserved positively charged residues. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9874.map.gz emd_9874.map.gz | 249.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9874-v30.xml emd-9874-v30.xml emd-9874.xml emd-9874.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9874_fsc.xml emd_9874_fsc.xml | 14.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_9874.png emd_9874.png | 32.7 KB | ||
| Filedesc metadata |  emd-9874.cif.gz emd-9874.cif.gz | 7.1 KB | ||
| Others |  emd_9874_additional_1.map.gz emd_9874_additional_1.map.gz emd_9874_additional_2.map.gz emd_9874_additional_2.map.gz | 212.5 MB 212.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9874 http://ftp.pdbj.org/pub/emdb/structures/EMD-9874 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9874 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9874 | HTTPS FTP |
-Related structure data
| Related structure data |  6jqmMC  9873C  9875C  9876C  6jqlC  6jqnC  6jqoC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9874.map.gz / Format: CCP4 / Size: 266.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9874.map.gz / Format: CCP4 / Size: 266.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This map is of PaaZ with NADPH bound. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
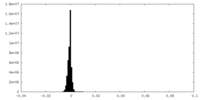
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: This map is one of the half-maps after refinement.
| File | emd_9874_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This map is one of the half-maps after refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
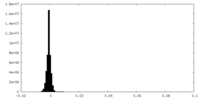
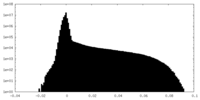
| Density Histograms |
-Additional map: This map is one of the half-maps after refinement.
| File | emd_9874_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This map is one of the half-maps after refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
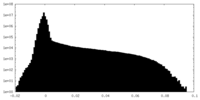
| Density Histograms |
- Sample components
Sample components
-Entire : PaaZ
| Entire | Name: PaaZ |
|---|---|
| Components |
|
-Supramolecule #1: PaaZ
| Supramolecule | Name: PaaZ / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: PaaZ is a bifunctional enzyme that has hydrolase and dehydrogenase activity. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 440 KDa |
-Macromolecule #1: Bifunctional protein PaaZ
| Macromolecule | Name: Bifunctional protein PaaZ / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: oxepin-CoA hydrolase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 73.969391 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHQQ LASFLSGTWQ SGRGRSRLIH HAISGEALWE VTSEGLDMAA ARQFAIEKGA PALRAMTFIE RAAMLKAVAK HLLSEKERF YALSAQTGAT RADSWVDIEG GIGTLFTYAS LGSRELPDDT LWPEDELIPL SKEGGFAARH LLTSKSGVAV H INAFNFPC ...String: MGHHHHHHQQ LASFLSGTWQ SGRGRSRLIH HAISGEALWE VTSEGLDMAA ARQFAIEKGA PALRAMTFIE RAAMLKAVAK HLLSEKERF YALSAQTGAT RADSWVDIEG GIGTLFTYAS LGSRELPDDT LWPEDELIPL SKEGGFAARH LLTSKSGVAV H INAFNFPC WGMLEKLAPT WLGGMPAIIK PATATAQLTQ AMVKSIVDSG LVPEGAISLI CGSAGDLLDH LDSQDVVTFT GS AATGQML RVQPNIVAKS IPFTMEADSL NCCVLGEDVT PDQPEFALFI REVVREMTTK AGQKCTAIRR IIVPQALVNA VSD ALVARL QKVVVGDPAQ EGVKMGALVN AEQRADVQEK VNILLAAGCE IRLGGQADLS AAGAFFPPTL LYCPQPDETP AVHA TEAFG PVATLMPAQN QRHALQLACA GGGSLAGTLV TADPQIARQF IADAARTHGR IQILNEESAK ESTGHGSPLP QLVHG GPGR AGGGEELGGL RAVKHYMQRT AVQGSPTMLA AISKQWVRGA KVEEDRIHPF RKYFEELQPG DSLLTPRRTM TEADIV NFA CLSGDHFYAH MDKIAAAESI FGERVVHGYF VLSAAAGLFV DAGVGPVIAN YGLESLRFIE PVKPGDTIQV RLTCKRK TL KKQRSAEEKP TGVVEWAVEV FNQHQTPVAL YSILTLVARQ HGDFVD UniProtKB: Bifunctional protein PaaZ |
-Macromolecule #2: NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE
| Macromolecule | Name: NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE type: ligand / ID: 2 / Number of copies: 6 / Formula: NDP |
|---|---|
| Molecular weight | Theoretical: 745.421 Da |
| Chemical component information |  ChemComp-NDP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.015 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 / Component:
Details: Protein was purified and kept in 25mM Hepes buffer and 50 mM NaCl | ||||||
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 300 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.02 kPa Details: Grids was glow discharged for 5 minutes and then graphene oxide was applied. The grids were washed with water 3 times and dried before using. | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV Details: blotting force 10, blotting time 4 sec, waiting time 15 sec, drying time 0, blotting times 1.. | ||||||
| Details | The peak fraction from gel filtration was used for grid preparation. 10-fold excess of NADPH was added before grid freezing. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 80.0 K / Max: 80.0 K |
| Details | Data was collected with EPU software |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 732 / Average exposure time: 60.0 sec. / Average electron dose: 27.0 e/Å2 Details: The 60 second exposure was saved into 75 frames with each frame ~0.36 e-. The frames were then grouped into 3 for alignment and summed images were used for data processing |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 132075 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 2.2 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Real space refinement with secondary structure enabled, minimization and adp |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 68.9 |
| Output model |  PDB-6jqm: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)