+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9774 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Capsid structure of a freshwater cyanophage Siphoviridae Mic1 | |||||||||||||||
 Map data Map data | the mrc map after postprocess | |||||||||||||||
 Sample Sample | Cyanophage != Microcystis phage Mic1 Cyanophage
| |||||||||||||||
 Keywords Keywords | cyanophage / Siphoviridae / capsid / VIRUS | |||||||||||||||
| Function / homology | : / Cement / Major capsid protein Function and homology information Function and homology information | |||||||||||||||
| Biological species |  Microcystis phage Mic1 (virus) Microcystis phage Mic1 (virus) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.53 Å | |||||||||||||||
 Authors Authors | Jin H / Jiang YL | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2019 Journal: Structure / Year: 2019Title: Capsid Structure of a Freshwater Cyanophage Siphoviridae Mic1. Authors: Hua Jin / Yong-Liang Jiang / Feng Yang / Jun-Tao Zhang / Wei-Fang Li / Ke Zhou / Jue Ju / Yuxing Chen / Cong-Zhao Zhou /  Abstract: Cyanobacteria are the most abundant photosynthetic microorganisms, the global distribution of which is mainly regulated by the corresponding cyanophages. A systematic screening of water samples in ...Cyanobacteria are the most abundant photosynthetic microorganisms, the global distribution of which is mainly regulated by the corresponding cyanophages. A systematic screening of water samples in the Lake Chaohu enabled us to isolate a freshwater siphocyanophage that infects Microcystis wesenbergii, thus termed Mic1. Using cryoelectron microscopy, we solved the 3.5-Å structure of Mic1 capsid. The major capsid protein gp40 of an HK97-like fold forms two types of capsomers, hexons and pentons. The capsomers interact with each other via the interweaved N-terminal arms of gp40 in addition to a tail-in-mouth joint along the three-fold symmetric axis, resulting in the assembly of capsid in a mortise-and-tenon pattern. The novel-fold cement protein gp47 sticks at the two-fold symmetric axis and further fixes the capsid. These findings provide structural insights into the assembly of cyanophages, and set up a platform to explore the mechanism of specific interactions and co-evolution with cyanobacteria. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9774.map.gz emd_9774.map.gz | 305 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9774-v30.xml emd-9774-v30.xml emd-9774.xml emd-9774.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9774.png emd_9774.png | 338.5 KB | ||
| Filedesc metadata |  emd-9774.cif.gz emd-9774.cif.gz | 5.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9774 http://ftp.pdbj.org/pub/emdb/structures/EMD-9774 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9774 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9774 | HTTPS FTP |
-Validation report
| Summary document |  emd_9774_validation.pdf.gz emd_9774_validation.pdf.gz | 559.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9774_full_validation.pdf.gz emd_9774_full_validation.pdf.gz | 559 KB | Display | |
| Data in XML |  emd_9774_validation.xml.gz emd_9774_validation.xml.gz | 9.8 KB | Display | |
| Data in CIF |  emd_9774_validation.cif.gz emd_9774_validation.cif.gz | 11.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9774 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9774 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9774 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9774 | HTTPS FTP |
-Related structure data
| Related structure data |  6j3qMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9774.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9774.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | the mrc map after postprocess | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
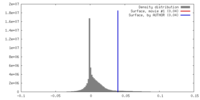
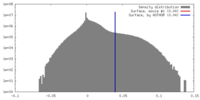
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cyanophage
| Entire | Name: Cyanophage |
|---|---|
| Components |
|
-Supramolecule #1: Microcystis phage Mic1
| Supramolecule | Name: Microcystis phage Mic1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2587456 / Sci species name: Microcystis phage Mic1 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Microcystis wesenbergii (bacteria) Microcystis wesenbergii (bacteria) |
| Virus shell | Shell ID: 1 / Name: gp40 / Diameter: 880.0 Å / T number (triangulation number): 13 |
-Macromolecule #1: major capsid protein
| Macromolecule | Name: major capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 13 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Microcystis phage Mic1 (virus) Microcystis phage Mic1 (virus) |
| Molecular weight | Theoretical: 37.879035 KDa |
| Sequence | String: MAKLNLAVLN NVFSELLTQE INRSATFLGL LSKYPERKSN IQWGVGMGGT TATGVAITGS APAASMDATL PAQLPISAAS VQSTFTLNL KEIEESKEQV TNEELRNLLE AQMRNAVEEI ATTLNKKLYD GSGAIADGGL IGLSIAASGQ DYAGISSATY P LWNVSEVD ...String: MAKLNLAVLN NVFSELLTQE INRSATFLGL LSKYPERKSN IQWGVGMGGT TATGVAITGS APAASMDATL PAQLPISAAS VQSTFTLNL KEIEESKEQV TNEELRNLLE AQMRNAVEEI ATTLNKKLYD GSGAIADGGL IGLSIAASGQ DYAGISSATY P LWNVSEVD AWDANATGTD KRQALKTDFM LELDRKIRYR PGAYDLILTT PKVVEQYKKV FEANRSYQIM TFDGQRVPLI DL GFNVAGY MGRPIIDDVF CSRTRTAAES AITTALGTAV DEGVMYFLKK DDLRFYSTPV VGAFSANGVY TLMRQLAQTS LYV DNFVVG CIPQLQLTTR KNVGVIKNIK VG |
-Macromolecule #2: cement protein
| Macromolecule | Name: cement protein / type: protein_or_peptide / ID: 2 / Number of copies: 13 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Microcystis phage Mic1 (virus) Microcystis phage Mic1 (virus) |
| Molecular weight | Theoretical: 10.199458 KDa |
| Sequence | String: MPLVYTPAVR GGANPASGSY LLDPQYVNSG VDILQATYGY NINGTANADQ LLQRDAILAI LEYALKDTAF VNAIQAVAAG SGVTTPASF VSACVTKLTA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 50 mM Tris-HCl pH7.5, 100 mM NaCl, 10 mM MgSO4 |
|---|---|
| Grid | Model: Quantifoil R3.5/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK I |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Number real images: 1935 / Average exposure time: 5.76 sec. / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6j3q: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)