+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9662 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 19A ZPC cryo-EM map of Eh V-ATPase-Fab | |||||||||
 Map data Map data | 19A ZPC cryo-EM map of Eh V-ATPase-Fab | |||||||||
 Sample Sample |
| |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 19.0 Å | |||||||||
 Authors Authors | Tsunoda J / Song C / Imai FL / Murata T / Ueno H / Iino R / Takagi J / Murata K | |||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2018 Journal: Sci Rep / Year: 2018Title: Off-axis rotor in Enterococcus hirae V-ATPase visualized by Zernike phase plate single-particle cryo-electron microscopy. Authors: Jun Tsunoda / Chihong Song / Fabiana Lica Imai / Junichi Takagi / Hiroshi Ueno / Takeshi Murata / Ryota Iino / Kazuyoshi Murata /  Abstract: EhV-ATPase is an ATP-driven Na pump in the eubacteria Enterococcus hirae (Eh). Here, we present the first entire structure of detergent-solubilized EhV-ATPase by single-particle cryo-electron ...EhV-ATPase is an ATP-driven Na pump in the eubacteria Enterococcus hirae (Eh). Here, we present the first entire structure of detergent-solubilized EhV-ATPase by single-particle cryo-electron microscopy (cryo-EM) using Zernike phase plate. The cryo-EM map dominantly showed one of three catalytic conformations in this rotary enzyme. To further stabilize the originally heterogeneous structure caused by the ATP hydrolysis states of the V-ATPases, a peptide epitope tag system was adopted, in which the inserted peptide epitope sequence interfered with rotation of the central rotor by binding the Fab. As a result, the map unexpectedly showed another catalytic conformation of EhV-ATPase. Interestingly, these two conformations identified with and without Fab conversely coincided with those of the minor state 2 and the major state 1 of Thermus thermophilus V/A-ATPase, respectively. The most prominent feature in EhV-ATPase was the off-axis rotor, where the cytoplasmic V domain was connected to the transmembrane V domain through the off-axis central rotor. Furthermore, compared to the structure of ATP synthases, the larger size of the interface between the transmembrane a-subunit and c-ring of EhV-ATPase would be more advantageous for active ion pumping. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9662.map.gz emd_9662.map.gz | 27.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9662-v30.xml emd-9662-v30.xml emd-9662.xml emd-9662.xml | 12.2 KB 12.2 KB | Display Display |  EMDB header EMDB header |
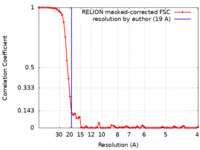
| FSC (resolution estimation) |  emd_9662_fsc.xml emd_9662_fsc.xml | 6.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_9662.png emd_9662.png | 46.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9662 http://ftp.pdbj.org/pub/emdb/structures/EMD-9662 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9662 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9662 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9662.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9662.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 19A ZPC cryo-EM map of Eh V-ATPase-Fab | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.992 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Enterococcus hirae V-ATPse
| Entire | Name: Enterococcus hirae V-ATPse |
|---|---|
| Components |
|
-Supramolecule #1: Enterococcus hirae V-ATPse
| Supramolecule | Name: Enterococcus hirae V-ATPse / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 800 kDa/nm |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Sugar embedding | Material: ice |
| Grid | Model: Quantifoil R1.2/1.3 / Material: MOLYBDENUM / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2200FS |
|---|---|
| Temperature | Min: 76.0 K / Max: 76.0 K |
| Specialist optics | Phase plate: ZERNIKE PHASE PLATE / Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 5120 pixel / Digitization - Dimensions - Height: 3840 pixel / Digitization - Sampling interval: 6.4 µm / Digitization - Frames/image: 3-75 / Number grids imaged: 5 / Number real images: 1302 / Average exposure time: 3.0 sec. / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 40.0 µm / Calibrated defocus max: 0.5 µm / Calibrated defocus min: 0.25 µm / Calibrated magnification: 32129 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 4.2 mm / Nominal defocus max: 0.5 µm / Nominal defocus min: 0.25 µm / Nominal magnification: 30000 |
| Sample stage | Specimen holder model: GATAN 914 HIGH TILT LIQUID NITROGEN CRYO TRANSFER TOMOGRAPHY HOLDER Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)