登録情報 データベース : EMDB / ID : EMD-9527タイトル Cryo-EM structure of the Catalytic Step I spliceosome (C complex) at 4.6 angstrom resolution Cryo-EM map of Catalytic Step I spliceosome (C complex) at 4.6 angstrom resolution 複合体 : Catalytic Step I spliceosome (C complex)機能・相同性 分子機能 ドメイン・相同性 構成要素
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Saccharomyces cerevisiae S288c (パン酵母)手法 / / 解像度 : 4.6 Å Wan R / Yan C / Bai R / Huang G / Shi Y ジャーナル : Science / 年 : 2016タイトル : Structure of a yeast catalytic step I spliceosome at 3.4 Å resolution.著者 : Ruixue Wan / Chuangye Yan / Rui Bai / Gaoxingyu Huang / Yigong Shi / 要旨 : Each cycle of pre-messenger RNA splicing, carried out by the spliceosome, comprises two sequential transesterification reactions, which result in the removal of an intron and the joining of two exons. ... Each cycle of pre-messenger RNA splicing, carried out by the spliceosome, comprises two sequential transesterification reactions, which result in the removal of an intron and the joining of two exons. Here we report an atomic structure of a catalytic step I spliceosome (known as the C complex) from Saccharomyces cerevisiae, as determined by cryo-electron microscopy at an average resolution of 3.4 angstroms. In the structure, the 2'-OH of the invariant adenine nucleotide in the branch point sequence (BPS) is covalently joined to the phosphate at the 5' end of the 5' splice site (5'SS), forming an intron lariat. The freed 5' exon remains anchored to loop I of U5 small nuclear RNA (snRNA), and the 5'SS and BPS of the intron form duplexes with conserved U6 and U2 snRNA sequences, respectively. Specific placement of these RNA elements at the catalytic cavity of Prp8 is stabilized by 15 protein components, including Snu114 and the splicing factors Cwc21, Cwc22, Cwc25, and Yju2. These features, representing the conformation of the spliceosome after the first-step reaction, predict structural changes that are needed for the execution of the second-step transesterification reaction. 履歴 登録 2016年7月14日 - ヘッダ(付随情報) 公開 2016年8月17日 - マップ公開 2016年8月17日 - 更新 2016年10月12日 - 現状 2016年10月12日 処理サイト : PDBj / 状態 : 公開
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
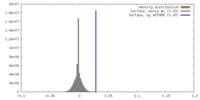
基本情報 マップデータ
マップデータ 試料
試料 機能・相同性情報
機能・相同性情報
 データ登録者
データ登録者 引用
引用 ジャーナル: Science / 年: 2016
ジャーナル: Science / 年: 2016
 構造の表示
構造の表示 ムービービューア
ムービービューア SurfView
SurfView Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク emd_9527.map.gz
emd_9527.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-9527-v30.xml
emd-9527-v30.xml emd-9527.xml
emd-9527.xml EMDBヘッダ
EMDBヘッダ emd_9527.png
emd_9527.png http://ftp.pdbj.org/pub/emdb/structures/EMD-9527
http://ftp.pdbj.org/pub/emdb/structures/EMD-9527 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9527
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9527 emd_9527_validation.pdf.gz
emd_9527_validation.pdf.gz EMDB検証レポート
EMDB検証レポート emd_9527_full_validation.pdf.gz
emd_9527_full_validation.pdf.gz emd_9527_validation.xml.gz
emd_9527_validation.xml.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9527
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9527 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9527
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9527 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_9527.map.gz / 形式: CCP4 / 大きさ: 244.1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_9527.map.gz / 形式: CCP4 / 大きさ: 244.1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) 試料の構成要素
試料の構成要素
 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN
 画像解析
画像解析 ムービー
ムービー コントローラー
コントローラー
































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)