[English] 日本語
 Yorodumi
Yorodumi- EMDB-9023: In situ structure of 96-nm repeat unit of Tetrahymena ciliary axo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9023 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ structure of 96-nm repeat unit of Tetrahymena ciliary axoneme by TYGRESS method | |||||||||
 Map data Map data | In situ structure of 96 nm repeat unit from intact Tetrahymena ciliary axoneme resolved by a newly developed TYGRESS method | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Tetrahymena thermophila CU428 (eukaryote) Tetrahymena thermophila CU428 (eukaryote) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 12.0 Å | |||||||||
 Authors Authors | Song K / Shang Z / Fu X / Lou X / Grigorieff N / Nicastro D | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Methods / Year: 2020 Journal: Nat Methods / Year: 2020Title: In situ structure determination at nanometer resolution using TYGRESS. Authors: Kangkang Song / Zhiguo Shang / Xiaofeng Fu / Xiaochu Lou / Nikolaus Grigorieff / Daniela Nicastro /  Abstract: The resolution of subtomogram averages calculated from cryo-electron tomograms (cryo-ET) of crowded cellular environments is often limited owing to signal loss in, and misalignment of, the ...The resolution of subtomogram averages calculated from cryo-electron tomograms (cryo-ET) of crowded cellular environments is often limited owing to signal loss in, and misalignment of, the subtomograms. By contrast, single-particle cryo-electron microscopy (SP-cryo-EM) routinely reaches near-atomic resolution of isolated complexes. We report a method called 'tomography-guided 3D reconstruction of subcellular structures' (TYGRESS) that is a hybrid of cryo-ET and SP-cryo-EM, and is able to achieve close-to-nanometer resolution of complexes inside crowded cellular environments. TYGRESS combines the advantages of SP-cryo-EM (images with good signal-to-noise ratio and contrast, as well as minimal radiation damage) and subtomogram averaging (three-dimensional alignment of macromolecules in a complex sample). Using TYGRESS, we determined the structure of the intact ciliary axoneme with up to resolution of 12 Å. These results reveal many structural details that were not visible by cryo-ET alone. TYGRESS is generally applicable to cellular complexes that are amenable to subtomogram averaging. #1:  Journal: biorXiv Journal: biorXivTitle: Structure of the ciliary axoneme at nanometer resolution reconstructed by TYGRESS Authors: Song K / Shang Z / Fu X / Lou X / Grigorieff N / Nicastro D | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9023.map.gz emd_9023.map.gz | 747.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9023-v30.xml emd-9023-v30.xml emd-9023.xml emd-9023.xml | 9.7 KB 9.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9023.png emd_9023.png | 112.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9023 http://ftp.pdbj.org/pub/emdb/structures/EMD-9023 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9023 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9023 | HTTPS FTP |
-Validation report
| Summary document |  emd_9023_validation.pdf.gz emd_9023_validation.pdf.gz | 78.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9023_full_validation.pdf.gz emd_9023_full_validation.pdf.gz | 78.1 KB | Display | |
| Data in XML |  emd_9023_validation.xml.gz emd_9023_validation.xml.gz | 492 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9023 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9023 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9023 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9023 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9023.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9023.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | In situ structure of 96 nm repeat unit from intact Tetrahymena ciliary axoneme resolved by a newly developed TYGRESS method | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.112 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
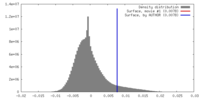
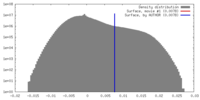
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : 96 nm repeat unit from Tetrahymena ciliary axoneme
| Entire | Name: 96 nm repeat unit from Tetrahymena ciliary axoneme |
|---|---|
| Components |
|
-Supramolecule #1: 96 nm repeat unit from Tetrahymena ciliary axoneme
| Supramolecule | Name: 96 nm repeat unit from Tetrahymena ciliary axoneme / type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Tetrahymena thermophila CU428 (eukaryote) / Organelle: Cilia Tetrahymena thermophila CU428 (eukaryote) / Organelle: Cilia |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.0 µm / Calibrated defocus min: 1.5 µm / Calibrated magnification: 9400 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 12.0 Å / Resolution method: FSC 0.143 CUT-OFF / Details: TYGRESS was used for the reconstruction method / Number images used: 18857 |
|---|---|
| Initial angle assignment | Type: OTHER / Details: From subtomogram alignment |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)