+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8851 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of B. subtilis flagellar filaments S17P | |||||||||
 Map data Map data | Cryo-EM structure of B. subtilis flagellar filaments S17P | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacteria flagella / helical polymers / cryo-EM / PROTEIN FIBRIL | |||||||||
| Function / homology | Flagellin, C-terminal domain / Bacterial flagellin C-terminal helical region / Flagellin / Flagellin, N-terminal domain / Bacterial flagellin N-terminal helical region / bacterial-type flagellum / structural molecule activity / extracellular region / Flagellin Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / cryo EM / negative staining / Resolution: 6.7 Å | |||||||||
 Authors Authors | Wang F / Burrage AM | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2017 Journal: Nat Commun / Year: 2017Title: A structural model of flagellar filament switching across multiple bacterial species. Authors: Fengbin Wang / Andrew M Burrage / Sandra Postel / Reece E Clark / Albina Orlova / Eric J Sundberg / Daniel B Kearns / Edward H Egelman /  Abstract: The bacterial flagellar filament has long been studied to understand how a polymer composed of a single protein can switch between different supercoiled states with high cooperativity. Here we ...The bacterial flagellar filament has long been studied to understand how a polymer composed of a single protein can switch between different supercoiled states with high cooperativity. Here we present near-atomic resolution cryo-EM structures for flagellar filaments from both Gram-positive Bacillus subtilis and Gram-negative Pseudomonas aeruginosa. Seven mutant flagellar filaments in B. subtilis and two in P. aeruginosa capture two different states of the filament. These reliable atomic models of both states reveal conserved molecular interactions in the interior of the filament among B. subtilis, P. aeruginosa and Salmonella enterica. Using the detailed information about the molecular interactions in two filament states, we successfully predict point mutations that shift the equilibrium between those two states. Further, we observe the dimerization of P. aeruginosa outer domains without any perturbation of the conserved interior of the filament. Our results give new insights into how the flagellin sequence has been "tuned" over evolution.Bacterial flagellar filaments are composed almost entirely of a single protein-flagellin-which can switch between different supercoiled states in a highly cooperative manner. Here the authors present near-atomic resolution cryo-EM structures of nine flagellar filaments, and begin to shed light on the molecular basis of filament switching. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8851.map.gz emd_8851.map.gz | 30.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8851-v30.xml emd-8851-v30.xml emd-8851.xml emd-8851.xml | 12.5 KB 12.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8851.png emd_8851.png | 107 KB | ||
| Filedesc metadata |  emd-8851.cif.gz emd-8851.cif.gz | 5.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8851 http://ftp.pdbj.org/pub/emdb/structures/EMD-8851 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8851 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8851 | HTTPS FTP |
-Related structure data
| Related structure data |  5wjxMC  8847C  8848C  8849C  8850C  8852C  8853C  8855C  8856C  5wjtC  5wjuC  5wjvC  5wjwC  5wjyC  5wjzC  5wk5C  5wk6C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8851.map.gz / Format: CCP4 / Size: 84.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8851.map.gz / Format: CCP4 / Size: 84.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of B. subtilis flagellar filaments S17P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
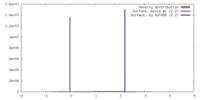
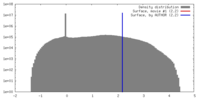
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Bacillus subtilis flagella filament
| Entire | Name: Bacillus subtilis flagella filament |
|---|---|
| Components |
|
-Supramolecule #1: Bacillus subtilis flagella filament
| Supramolecule | Name: Bacillus subtilis flagella filament / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Flagellin
| Macromolecule | Name: Flagellin / type: protein_or_peptide / ID: 1 / Number of copies: 46 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 32.67132 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRINHNIAAL NTLNRLPSNN SASQKNMEKL SSGLRINRAG DDAAGLAISE KMRGQIRGLE MASKNSQDGI SLIQTAEGAL TETHAILQR VRELVVQAGN TGTQDKATDL QSIQDEISAL TDEIDGISNR TEFNGKKLLD GTYKVDTATP ANQKNLVFQI G ANATQQIS ...String: MRINHNIAAL NTLNRLPSNN SASQKNMEKL SSGLRINRAG DDAAGLAISE KMRGQIRGLE MASKNSQDGI SLIQTAEGAL TETHAILQR VRELVVQAGN TGTQDKATDL QSIQDEISAL TDEIDGISNR TEFNGKKLLD GTYKVDTATP ANQKNLVFQI G ANATQQIS VNIEDMGADA LGIKEADGSI AALHSVNDLD VTKFADNAAD CADIGFDAQL KVVDEAINQV SSQRAKLGAV QN RLEHTIN NLSASGENLT AAESRIRDVD MAKEMSEFTK NNILSQASQA MLAQANQQPQ NVLQLLR UniProtKB: Flagellin |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 6.8 / Details: Imidazole buffer |
| Staining | Type: NEGATIVE / Material: negative stain |
| Grid | Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Average exposure time: 2.0 sec. / Average electron dose: 20.0 e/Å2 Details: Images were stored containing seven parts, where each part represented a set of frames corresponding to a dose of ~20 electrons per Angstrom^2. The full dose image stack was used for the ...Details: Images were stored containing seven parts, where each part represented a set of frames corresponding to a dose of ~20 electrons per Angstrom^2. The full dose image stack was used for the estimation of the CTF as well as for boxing filaments. Only the first two parts were used for the reconstruction (~5 electrons per Angstrom^2). |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 4.68 Å Applied symmetry - Helical parameters - Δ&Phi: 65.29 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Algorithm: BACK PROJECTION / Resolution.type: BY AUTHOR / Resolution: 6.7 Å / Resolution method: OTHER / Software - Name: SPIDER / Details: model-map FSC 0.38 cut-off / Number images used: 13899 |
|---|---|
| Startup model | Type of model: OTHER / Details: featureless cylinder |
| Final angle assignment | Type: NOT APPLICABLE / Software - Name: SPIDER |
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-5wjx: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)