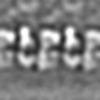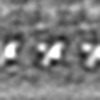+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8835 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Sub-tomogram average of ODA Post class | |||||||||
 Map data Map data | Sub-tomogram average of ODA Post class: sea urchin sperm flagellar ODA dyneins are in post-powerstroke state | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 33.0 Å | |||||||||
 Authors Authors | Lin J / Nicastro D | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2018 Journal: Science / Year: 2018Title: Asymmetric distribution and spatial switching of dynein activity generates ciliary motility. Authors: Jianfeng Lin / Daniela Nicastro /  Abstract: Motile cilia and flagella are essential, highly conserved organelles, and their motility is driven by the coordinated activities of multiple dynein isoforms. The prevailing "switch-point" hypothesis ...Motile cilia and flagella are essential, highly conserved organelles, and their motility is driven by the coordinated activities of multiple dynein isoforms. The prevailing "switch-point" hypothesis posits that dyneins are asymmetrically activated to drive flagellar bending. To test this model, we applied cryo-electron tomography to visualize activity states of individual dyneins relative to their locations along beating flagella of sea urchin sperm cells. As predicted, bending was generated by the asymmetric distribution of dynein activity on opposite sides of the flagellum. However, contrary to predictions, most dyneins were in their active state, and the smaller population of conformationally inactive dyneins switched flagellar sides relative to the bending direction. Thus, our data suggest a "switch-inhibition" mechanism in which force imbalance is generated by inhibiting, rather than activating, dyneins on alternating sides of the flagellum. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8835.map.gz emd_8835.map.gz | 566.1 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8835-v30.xml emd-8835-v30.xml emd-8835.xml emd-8835.xml | 11.4 KB 11.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8835.png emd_8835.png | 82.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8835 http://ftp.pdbj.org/pub/emdb/structures/EMD-8835 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8835 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8835 | HTTPS FTP |
-Related structure data
| Related structure data |  8836C  8837C  8838C C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10158 (Title: Cryo electron tomography of immotile sea urchin sperm flagella EMPIAR-10158 (Title: Cryo electron tomography of immotile sea urchin sperm flagellaData size: 13.8 Data #1: Tilt series of immotile sea urchin sperm flagella [tilt series]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8835.map.gz / Format: CCP4 / Size: 686.5 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8835.map.gz / Format: CCP4 / Size: 686.5 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sub-tomogram average of ODA Post class: sea urchin sperm flagellar ODA dyneins are in post-powerstroke state | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 9.856 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
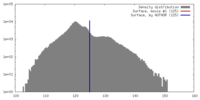
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Sea urchin sperm
| Entire | Name: Sea urchin sperm |
|---|---|
| Components |
|
-Supramolecule #1: Sea urchin sperm
| Supramolecule | Name: Sea urchin sperm / type: cell / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Sperm cells were treated with ATPase inhibitor erythro-9-(2-Hydroxy-3-nonyl)adenine hydrochloride (EHNA hydrochloride, 2 mM). |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Specialist optics | Energy filter - Name: GIF |
| Image recording | Film or detector model: GATAN MULTISCAN / Average electron dose: 1.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 8.0 µm / Nominal defocus min: 6.0 µm / Nominal magnification: 13500 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 33.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: PEET (ver. 1.9.0) / Number subtomograms used: 2000 |
|---|---|
| Extraction | Number tomograms: 20 / Number images used: 14066 / Software - Name: PEET (ver. 1.9.0) |
| Final 3D classification | Software - Name: PEET (ver. 1.9.0) |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)