[English] 日本語
 Yorodumi
Yorodumi- EMDB-8801: Negative-stain EM reconstruction of H3v-47 Fab in complex with A/... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8801 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative-stain EM reconstruction of H3v-47 Fab in complex with A/Minnesota/11/2010 H3N2v HA and FI6v Fab | |||||||||
 Map data Map data | Reconstruction of HA in complex with H3v-47 Fab and FI6v Fab. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Influenza A virus / Influenza A virus /  Homo sapiens (human) / Homo sapiens (human) /  Human (human) Human (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 25.8 Å | |||||||||
 Authors Authors | Nieusma T / Ward AB | |||||||||
 Citation Citation |  Journal: JCI Insight / Year: 2016 Journal: JCI Insight / Year: 2016Title: Recognition of influenza H3N2 variant virus by human neutralizing antibodies. Authors: Sandhya Bangaru / Travis Nieusma / Nurgun Kose / Natalie J Thornburg / Jessica A Finn / Bryan S Kaplan / Hannah G King / Vidisha Singh / Rebecca M Lampley / Gopal Sapparapu / Alberto ...Authors: Sandhya Bangaru / Travis Nieusma / Nurgun Kose / Natalie J Thornburg / Jessica A Finn / Bryan S Kaplan / Hannah G King / Vidisha Singh / Rebecca M Lampley / Gopal Sapparapu / Alberto Cisneros / Kathryn M Edwards / James C Slaughter / Srilatha Edupuganti / Lilin Lai / Juergen A Richt / Richard J Webby / Andrew B Ward / James E Crowe /  Abstract: Since 2011, over 300 human cases of infection, especially in exposed children, with the influenza A H3N2 variant (H3N2v) virus that circulates in swine in the US have been reported. The structural ...Since 2011, over 300 human cases of infection, especially in exposed children, with the influenza A H3N2 variant (H3N2v) virus that circulates in swine in the US have been reported. The structural and genetic basis for the lack of protection against H3N2v induced by vaccines containing seasonal H3N2 antigens is poorly understood. We isolated 17 human monoclonal antibodies (mAbs) that neutralized H3N2v virus from subjects experimentally immunized with an H3N2v candidate vaccine. Six mAbs exhibited very potent neutralizing activity (IC < 200 ng/ml) against the H3N2v virus but not against current human H3N2 circulating strains. Fine epitope mapping and structural characterization of antigen-antibody complexes revealed that H3N2v specificity was attributable to amino acid polymorphisms in the 150-loop and the 190-helix antigenic sites on the hemagglutinin protein. H3N2v-specific antibodies also neutralized human H3N2 influenza strains naturally circulating between 1995 and 2005. These results reveal a high level of antigenic relatedness between the swine H3N2v virus and previously circulating human strains, consistent with the fact that early human H3 seasonal strains entered the porcine population in the 1990s and reentered the human population, where they had not been circulating, as H3N2v about a decade later. The data also explain the increased susceptibility to H3N2v viruses in young children, who lack prior exposure to human seasonal strains from the 1990s. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8801.map.gz emd_8801.map.gz | 13.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8801-v30.xml emd-8801-v30.xml emd-8801.xml emd-8801.xml | 11.6 KB 11.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8801.png emd_8801.png | 58.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8801 http://ftp.pdbj.org/pub/emdb/structures/EMD-8801 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8801 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8801 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8801.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8801.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of HA in complex with H3v-47 Fab and FI6v Fab. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
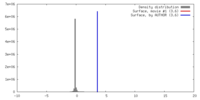
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : A/Minnesota/11/2010 H3N2v HA
| Entire | Name: A/Minnesota/11/2010 H3N2v HA |
|---|---|
| Components |
|
-Supramolecule #1: A/Minnesota/11/2010 H3N2v HA
| Supramolecule | Name: A/Minnesota/11/2010 H3N2v HA / type: complex / ID: 1 / Parent: 0 Details: H3v-47 Fab in complex with A/Minnesota/11/2010 H3N2v HA and FI6v Fab |
|---|---|
| Source (natural) | Organism:   Influenza A virus / Strain: H3N2 Influenza A virus / Strain: H3N2 |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK Freestyle 293-F / Recombinant plasmid: pcDNA3.1(+) Homo sapiens (human) / Recombinant cell: HEK Freestyle 293-F / Recombinant plasmid: pcDNA3.1(+) |
| Molecular weight | Theoretical: 50 KDa |
-Supramolecule #2: H3v-47 Fab
| Supramolecule | Name: H3v-47 Fab / type: complex / ID: 2 / Parent: 1 Details: H3v-47 Fab in complex with A/Minnesota/11/2010 H3N2v HA and FI6v Fab |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: human hybridoma cell line Homo sapiens (human) / Recombinant cell: human hybridoma cell line |
-Supramolecule #3: FI6v Fab
| Supramolecule | Name: FI6v Fab / type: complex / ID: 3 / Parent: 1 Details: Negative-stain EM reconstruction of H3v-47 Fab in complex with A/Minnesota/11/2010 H3N2v HA and FI6v Fab |
|---|---|
| Source (natural) | Organism:  Human (human) / Strain: H3N2 Human (human) / Strain: H3N2 |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl formate Details: The complex was diluted to 2ug/ml, applied to freshly glow discharged 400 mesh carbon coated copper grids, and negatively stained with 2% uranyl formate. |
| Grid | Model: EMS / Material: COPPER / Mesh: 400 / Support film - Material: CELLULOSE ACETATE / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Details: Carbon was evaporated upon grid prior to use. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Average electron dose: 24.7 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 1.0 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 52000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 20.0 mm / Nominal defocus max: 1.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 52000 |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 25.8 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN (ver. 1.1) / Number images used: 23175 |
|---|---|
| Initial angle assignment | Type: COMMON LINE |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera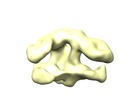






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)