+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8778 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
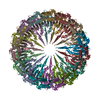
| Title | Type II secretin from Enteropathogenic Escherichia coli - GspD | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Type 2 secretin / outer membrane complex / homo oligomer / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein secretion by the type II secretion system / type II protein secretion system complex / cell outer membrane / identical protein binding Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Hay ID / Belousoff MJ | |||||||||
| Funding support |  Australia, 2 items Australia, 2 items
| |||||||||
 Citation Citation |  Journal: J Bacteriol / Year: 2018 Journal: J Bacteriol / Year: 2018Title: Structure and Membrane Topography of the Vibrio-Type Secretin Complex from the Type 2 Secretion System of Enteropathogenic Escherichia coli. Authors: Iain D Hay / Matthew J Belousoff / Rhys A Dunstan / Rebecca S Bamert / Trevor Lithgow /  Abstract: The β-barrel assembly machinery (BAM) complex is the core machinery for the assembly of β-barrel membrane proteins, and inhibition of BAM complex activity is lethal to bacteria. Discovery of ...The β-barrel assembly machinery (BAM) complex is the core machinery for the assembly of β-barrel membrane proteins, and inhibition of BAM complex activity is lethal to bacteria. Discovery of integral membrane proteins that are key to pathogenesis and yet do not require assistance from the BAM complex raises the question of how these proteins assemble into bacterial outer membranes. Here, we address this question through a structural analysis of the type 2 secretion system (T2SS) secretin from enteropathogenic O127:H6 strain E2348/69. Long β-strands assemble into a barrel extending 17 Å through and beyond the outer membrane, adding insight to how these extensive β-strands are assembled into the outer membrane. The substrate docking chamber of this secretin is shown to be sufficient to accommodate the substrate mucinase SteC. In order to cause disease, bacterial pathogens inhibit immune responses and induce pathology that will favor their replication and dissemination. In Gram-negative bacteria, these key attributes of pathogenesis depend on structures assembled into or onto the outer membrane. One of these is the T2SS. The -type T2SS mediates cholera toxin secretion in , and in O127:H6 strain E2348/69, the same machinery mediates secretion of the mucinases that enable the pathogen to penetrate intestinal mucus and thereby establish deadly infections. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8778.map.gz emd_8778.map.gz | 85.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8778-v30.xml emd-8778-v30.xml emd-8778.xml emd-8778.xml | 12.9 KB 12.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8778.png emd_8778.png | 54.9 KB | ||
| Filedesc metadata |  emd-8778.cif.gz emd-8778.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8778 http://ftp.pdbj.org/pub/emdb/structures/EMD-8778 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8778 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8778 | HTTPS FTP |
-Validation report
| Summary document |  emd_8778_validation.pdf.gz emd_8778_validation.pdf.gz | 530.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8778_full_validation.pdf.gz emd_8778_full_validation.pdf.gz | 530.1 KB | Display | |
| Data in XML |  emd_8778_validation.xml.gz emd_8778_validation.xml.gz | 6.5 KB | Display | |
| Data in CIF |  emd_8778_validation.cif.gz emd_8778_validation.cif.gz | 7.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8778 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8778 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8778 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8778 | HTTPS FTP |
-Related structure data
| Related structure data |  5w68MC  8779C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8778.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8778.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Type II sectretin outer membrane complex
| Entire | Name: Type II sectretin outer membrane complex |
|---|---|
| Components |
|
-Supramolecule #1: Type II sectretin outer membrane complex
| Supramolecule | Name: Type II sectretin outer membrane complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Putative type II secretion protein
| Macromolecule | Name: Putative type II secretion protein / type: protein_or_peptide / ID: 1 / Number of copies: 15 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: E2348/69 / EPEC |
| Molecular weight | Theoretical: 41.192559 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GNSQVFYLKY SKAEDLVDVL KQVSGTLTAA KEEAEGTVGS GREVVSIAAS KHSNALIVTA PQDIMQSLQS VIEQLDIRRA QVHVEALIV EVAEGSNINF GVQWGSKDAG LMQFANGTQI PIGTLGAAIS AAKPQKGSTV ISENGATTIN PDTNGDLSTL A QLLSGFSG ...String: GNSQVFYLKY SKAEDLVDVL KQVSGTLTAA KEEAEGTVGS GREVVSIAAS KHSNALIVTA PQDIMQSLQS VIEQLDIRRA QVHVEALIV EVAEGSNINF GVQWGSKDAG LMQFANGTQI PIGTLGAAIS AAKPQKGSTV ISENGATTIN PDTNGDLSTL A QLLSGFSG TAVGVVKGDW MALVQAVKND SSSNVLSTPS ITTLDNQEAF FMVGQDVPVL TGSTVGSNNS NPFNTVERKK VG IMLKVTP QINEGNAVQM VIEQEVSKVE GQTSLDVVFG ERKLKTTVLA NDGELIVLGG LMDDQAGESV AKVPLLGDIP LIG NLFKST ADKKEKRNLM VFIRPTILRD GMAADGVSQR KYNYMRAEQI YRDEQGLSLM PHTAQPILPA Q UniProtKB: Putative type II secretion protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-18 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 130000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-5w68: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















