+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8357 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Autoinhibited E. coli ATP synthase state 1 | |||||||||
 Map data Map data | Autoinhibited E. coli ATP synthase state 1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ATP synthase / ATPase / rotary motor / membrane protein / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationproton motive force-driven plasma membrane ATP synthesis / proton motive force-driven ATP synthesis / proton-transporting two-sector ATPase complex, proton-transporting domain / proton-transporting ATPase activity, rotational mechanism / H+-transporting two-sector ATPase / proton-transporting ATP synthase complex / proton-transporting ATP synthase activity, rotational mechanism / ADP binding / lipid binding / ATP hydrolysis activity ...proton motive force-driven plasma membrane ATP synthesis / proton motive force-driven ATP synthesis / proton-transporting two-sector ATPase complex, proton-transporting domain / proton-transporting ATPase activity, rotational mechanism / H+-transporting two-sector ATPase / proton-transporting ATP synthase complex / proton-transporting ATP synthase activity, rotational mechanism / ADP binding / lipid binding / ATP hydrolysis activity / ATP binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
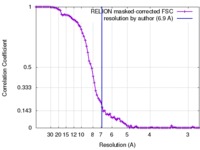
| Method | single particle reconstruction / cryo EM / Resolution: 6.9 Å | |||||||||
 Authors Authors | Sobti M / Smits C / Wong AS / Ishmukhametov R / Stock D / Sandin S / Stewart AG | |||||||||
 Citation Citation |  Journal: Elife / Year: 2016 Journal: Elife / Year: 2016Title: Cryo-EM structures of the autoinhibited ATP synthase in three rotational states. Authors: Meghna Sobti / Callum Smits / Andrew Sw Wong / Robert Ishmukhametov / Daniela Stock / Sara Sandin / Alastair G Stewart /    Abstract: A molecular model that provides a framework for interpreting the wealth of functional information obtained on the F-ATP synthase has been generated using cryo-electron microscopy. Three different ...A molecular model that provides a framework for interpreting the wealth of functional information obtained on the F-ATP synthase has been generated using cryo-electron microscopy. Three different states that relate to rotation of the enzyme were observed, with the central stalk's ε subunit in an extended autoinhibitory conformation in all three states. The F motor comprises of seven transmembrane helices and a decameric c-ring and invaginations on either side of the membrane indicate the entry and exit channels for protons. The proton translocating subunit contains near parallel helices inclined by ~30° to the membrane, a feature now synonymous with rotary ATPases. For the first time in this rotary ATPase subtype, the peripheral stalk is resolved over its entire length of the complex, revealing the F attachment points and a coiled-coil that bifurcates toward the membrane with its helices separating to embrace subunit from two sides. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8357.map.gz emd_8357.map.gz | 55.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8357-v30.xml emd-8357-v30.xml emd-8357.xml emd-8357.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8357_fsc.xml emd_8357_fsc.xml | 8.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_8357.png emd_8357.png | 45.3 KB | ||
| Filedesc metadata |  emd-8357.cif.gz emd-8357.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8357 http://ftp.pdbj.org/pub/emdb/structures/EMD-8357 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8357 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8357 | HTTPS FTP |
-Related structure data
| Related structure data |  5t4oMC  8358C  8359C  5t4pC  5t4qC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8357.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8357.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Autoinhibited E. coli ATP synthase state 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : ATP synthase
+Supramolecule #1: ATP synthase
+Macromolecule #1: ATP synthase subunit alpha
+Macromolecule #2: ATP synthase subunit beta
+Macromolecule #3: ATP synthase gamma chain
+Macromolecule #4: ATP synthase epsilon chain
+Macromolecule #5: ATP synthase subunit b
+Macromolecule #6: ATP synthase subunit a
+Macromolecule #7: ATP synthase subunit delta
+Macromolecule #8: ATP synthase subunit c
+Macromolecule #9: ADENOSINE-5'-TRIPHOSPHATE
+Macromolecule #10: ADENOSINE-5'-DIPHOSPHATE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 29.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)