[English] 日本語
 Yorodumi
Yorodumi- EMDB-8224: Structure of a mutant type IV pilus machinery (delta N1 ring dele... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8224 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a mutant type IV pilus machinery (delta N1 ring deletion in PilQ) in the closed state | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 50.0 Å | |||||||||
 Authors Authors | Gold VAM / Salzer R / Averhoff B | |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2016 Journal: J Biol Chem / Year: 2016Title: Topology and Structure/Function Correlation of Ring- and Gate-forming Domains in the Dynamic Secretin Complex of Thermus thermophilus. Authors: Ralf Salzer / Edoardo D'Imprima / Vicki A M Gold / Ilona Rose / Moritz Drechsler / Janet Vonck / Beate Averhoff /  Abstract: Secretins are versatile outer membrane pores used by many bacteria to secrete proteins, toxins, or filamentous phages; extrude type IV pili (T4P); or take up DNA. Extrusion of T4P and natural ...Secretins are versatile outer membrane pores used by many bacteria to secrete proteins, toxins, or filamentous phages; extrude type IV pili (T4P); or take up DNA. Extrusion of T4P and natural transformation of DNA in the thermophilic bacterium Thermus thermophilus requires a unique secretin complex comprising six stacked rings, a membrane-embedded cone structure, and two gates that open and close a central channel. To investigate the role of distinct domains in ring and gate formation, we examined a set of deletion derivatives by cryomicroscopy techniques. Here we report that maintaining the N0 ring in the deletion derivatives led to stable PilQ complexes. Analyses of the variants unraveled that an N-terminal domain comprising a unique βββαβ fold is essential for the formation of gate 2. Furthermore, we identified four βαββα domains essential for the formation of the N2 to N5 rings. Mutant studies revealed that deletion of individual ring domains significantly reduces piliation. The N1, N2, N4, and N5 deletion mutants were significantly impaired in T4P-mediated twitching motility, whereas the motility of the N3 mutant was comparable with that of wild-type cells. This indicates that the deletion of the N3 ring leads to increased pilus dynamics, thereby compensating for the reduced number of pili of the N3 mutant. All mutants exhibit a wild-type natural transformation phenotype, leading to the conclusion that DNA uptake is independent of functional T4P. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8224.map.gz emd_8224.map.gz | 355 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8224-v30.xml emd-8224-v30.xml emd-8224.xml emd-8224.xml | 10.6 KB 10.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8224.png emd_8224.png | 56.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8224 http://ftp.pdbj.org/pub/emdb/structures/EMD-8224 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8224 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8224 | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8224.map.gz / Format: CCP4 / Size: 501 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8224.map.gz / Format: CCP4 / Size: 501 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 11.56 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
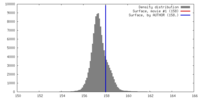
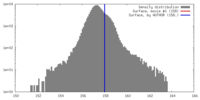
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Structure of a mutant type IV pilus machinery (delta N1 ring dele...
| Entire | Name: Structure of a mutant type IV pilus machinery (delta N1 ring deletion in PilQ) in the closed state from Thermus thermophilus |
|---|---|
| Components |
|
-Supramolecule #1: Structure of a mutant type IV pilus machinery (delta N1 ring dele...
| Supramolecule | Name: Structure of a mutant type IV pilus machinery (delta N1 ring deletion in PilQ) in the closed state from Thermus thermophilus type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus (bacteria) / Strain: HB27 Thermus thermophilus (bacteria) / Strain: HB27 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Component:
| ||||||
|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 20 % / Instrument: HOMEMADE PLUNGER Details: Cell solutions were mixed 1:1 (v/v) with 10 nm ProteinA-gold particles. 3 microlitres of sample were applied to grids and blotted on one side for ~5s before plunging. | ||||||
| Details | Cellular suspension |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 3.0 e/Å2 Details: Each image in every tilt series is a drift corrected sum of 3-5 frames |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 7.0 µm / Nominal magnification: 10000 |
| Sample stage | Specimen holder model: JEOL 3200FSC CRYOHOLDER / Cooling holder cryogen: NITROGEN |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C12 (12 fold cyclic) / Algorithm: BACK PROJECTION / Resolution.type: BY AUTHOR / Resolution: 50.0 Å / Resolution method: FSC 0.5 CUT-OFF Details: Resolution estimate was calculated using a mask to exclude the membrane and peptidoglycan. C12 symmetry was applied to the masked central PilQ core by rotation of each subvolume by 30 ...Details: Resolution estimate was calculated using a mask to exclude the membrane and peptidoglycan. C12 symmetry was applied to the masked central PilQ core by rotation of each subvolume by 30 degrees (360 degrees/12) prior to alignment search. Number subtomograms used: 432 |
|---|---|
| Extraction | Number tomograms: 8 / Number images used: 36 / Software - Name:  IMOD (ver. 4.8.46) IMOD (ver. 4.8.46) |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)