[English] 日本語
 Yorodumi
Yorodumi- EMDB-8149: Cryo-EM structure of a full archaeal ribosomal translation initia... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8149 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
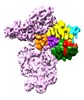
| Title | Cryo-EM structure of a full archaeal ribosomal translation initiation complex in the P-INconformation | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transcription | |||||||||
| Function / homology |  Function and homology information Function and homology informationtranslation reinitiation / formation of translation preinitiation complex / ribonuclease P activity / tRNA 5'-leader removal / protein-synthesizing GTPase / translation elongation factor activity / translation initiation factor activity / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA / translational initiation ...translation reinitiation / formation of translation preinitiation complex / ribonuclease P activity / tRNA 5'-leader removal / protein-synthesizing GTPase / translation elongation factor activity / translation initiation factor activity / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA / translational initiation / regulation of translation / ribosome biogenesis / ribosome binding / ribosomal small subunit assembly / ribosomal small subunit biogenesis / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / cytoplasmic translation / tRNA binding / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / GTPase activity / GTP binding / RNA binding / zinc ion binding / metal ion binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |   Pyrococcus abyssi GE5 (archaea) / Pyrococcus abyssi GE5 (archaea) /    Pyrococcus (archaea) Pyrococcus (archaea) | |||||||||
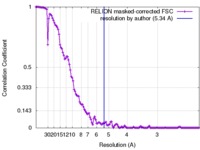
| Method | single particle reconstruction / cryo EM / Resolution: 5.34 Å | |||||||||
 Authors Authors | COUREUX P-D / SCHMITT E | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2016 Journal: Nat Commun / Year: 2016Title: Cryo-EM study of start codon selection during archaeal translation initiation. Authors: Pierre-Damien Coureux / Christine Lazennec-Schurdevin / Auriane Monestier / Eric Larquet / Lionel Cladière / Bruno P Klaholz / Emmanuelle Schmitt / Yves Mechulam /  Abstract: Eukaryotic and archaeal translation initiation complexes have a common structural core comprising e/aIF1, e/aIF1A, the ternary complex (TC, e/aIF2-GTP-Met-tRNA) and mRNA bound to the small ribosomal ...Eukaryotic and archaeal translation initiation complexes have a common structural core comprising e/aIF1, e/aIF1A, the ternary complex (TC, e/aIF2-GTP-Met-tRNA) and mRNA bound to the small ribosomal subunit. e/aIF2 plays a crucial role in this process but how this factor controls start codon selection remains unclear. Here, we present cryo-EM structures of the full archaeal 30S initiation complex showing two conformational states of the TC. In the first state, the TC is bound to the ribosome in a relaxed conformation with the tRNA oriented out of the P site. In the second state, the tRNA is accommodated within the peptidyl (P) site and the TC becomes constrained. This constraint is compensated by codon/anticodon base pairing, whereas in the absence of a start codon, aIF2 contributes to swing out the tRNA. This spring force concept highlights a mechanism of codon/anticodon probing by the initiator tRNA directly assisted by aIF2. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8149.map.gz emd_8149.map.gz | 153.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8149-v30.xml emd-8149-v30.xml emd-8149.xml emd-8149.xml | 59.9 KB 59.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8149_fsc.xml emd_8149_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_8149.png emd_8149.png | 124.6 KB | ||
| Filedesc metadata |  emd-8149.cif.gz emd-8149.cif.gz | 12.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8149 http://ftp.pdbj.org/pub/emdb/structures/EMD-8149 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8149 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8149 | HTTPS FTP |
-Related structure data
| Related structure data |  5jbhMC  8148C  5jb3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8149.map.gz / Format: CCP4 / Size: 160.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8149.map.gz / Format: CCP4 / Size: 160.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.12 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : 30S archaeal translation initiation complex
+Supramolecule #1: 30S archaeal translation initiation complex
+Supramolecule #2: 30S archaeal translation initiation complex
+Supramolecule #3: 30S archaeal translation initiation complex
+Supramolecule #4: 30S archaeal translation initiation complex
+Supramolecule #5: 30S archaeal translation initiation complex
+Macromolecule #1: 16S ribosomal RNA
+Macromolecule #30: mRNA
+Macromolecule #32: initiator Met-tRNA fMet from E. coli (A1U72 variant)
+Macromolecule #2: 30S ribosomal protein uS3
+Macromolecule #3: 50S ribosomal protein uL30
+Macromolecule #4: 30S ribosomal protein uS10
+Macromolecule #5: 30S ribosomal protein uS13
+Macromolecule #6: 30S ribosomal protein uS14
+Macromolecule #7: 30S ribosomal protein eS17
+Macromolecule #8: 30S ribosomal protein uS19
+Macromolecule #9: 30S ribosomal protein eS19
+Macromolecule #10: 30S ribosomal protein eS28
+Macromolecule #11: 30S ribosomal protein eS27
+Macromolecule #12: 30S ribosomal protein uS7
+Macromolecule #13: 30S ribosomal protein uS9
+Macromolecule #14: 30S ribosomal protein uS11
+Macromolecule #15: 30S ribosomal protein uS12
+Macromolecule #16: 30S ribosomal protein uS15
+Macromolecule #17: 30S ribosomal protein uS17
+Macromolecule #18: 30S ribosomal protein uS3
+Macromolecule #19: 30S ribosomal protein uS2
+Macromolecule #20: 30S ribosomal protein eS24
+Macromolecule #21: 30S ribosomal protein eS27
+Macromolecule #22: 30S ribosomal protein uS4
+Macromolecule #23: 30S ribosomal protein eS4
+Macromolecule #24: 30S ribosomal protein uS5
+Macromolecule #25: 30S ribosomal protein eS6
+Macromolecule #26: 30S ribosomal protein uS8
+Macromolecule #27: 30S ribosomal protein eS8
+Macromolecule #28: 30S ribosomal protein SX
+Macromolecule #29: 30S ribosomal protein eL41
+Macromolecule #31: aIF1
+Macromolecule #33: aIF1A
+Macromolecule #34: aIF2-gamma
+Macromolecule #35: aIF2-beta
+Macromolecule #36: aIF2-alpha
+Macromolecule #37: METHIONINE
+Macromolecule #38: MAGNESIUM ION
+Macromolecule #39: PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER
+Macromolecule #40: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 44.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)