+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7967 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Calmodulin-bound full-length rbTRPV5 | |||||||||
 Map data Map data | Calmodulin-bound rbTRPV5 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | calmodulin / TRPV5 / full-length / calcium channel / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of urine volume / : / type 3 metabotropic glutamate receptor binding / establishment of protein localization to membrane / negative regulation of ryanodine-sensitive calcium-release channel activity / organelle localization by membrane tethering / mitochondrion-endoplasmic reticulum membrane tethering / autophagosome membrane docking / negative regulation of calcium ion export across plasma membrane / regulation of cardiac muscle cell action potential ...regulation of urine volume / : / type 3 metabotropic glutamate receptor binding / establishment of protein localization to membrane / negative regulation of ryanodine-sensitive calcium-release channel activity / organelle localization by membrane tethering / mitochondrion-endoplasmic reticulum membrane tethering / autophagosome membrane docking / negative regulation of calcium ion export across plasma membrane / regulation of cardiac muscle cell action potential / nitric-oxide synthase binding / regulation of synaptic vesicle exocytosis / presynaptic endocytosis / calcineurin-mediated signaling / calcium ion import across plasma membrane / adenylate cyclase binding / regulation of ryanodine-sensitive calcium-release channel activity / protein phosphatase activator activity / regulation of synaptic vesicle endocytosis / detection of calcium ion / regulation of cardiac muscle contraction / postsynaptic cytosol / catalytic complex / phosphatidylinositol 3-kinase binding / calcium ion homeostasis / calcium channel inhibitor activity / presynaptic cytosol / cellular response to interferon-beta / regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / titin binding / regulation of calcium-mediated signaling / voltage-gated potassium channel complex / calcium channel complex / regulation of heart rate / calyx of Held / nitric-oxide synthase regulator activity / adenylate cyclase activator activity / response to amphetamine / sarcomere / protein serine/threonine kinase activator activity / regulation of cytokinesis / spindle microtubule / positive regulation of receptor signaling pathway via JAK-STAT / calcium channel regulator activity / response to calcium ion / calcium ion transmembrane transport / cellular response to type II interferon / G2/M transition of mitotic cell cycle / Schaffer collateral - CA1 synapse / calcium channel activity / spindle pole / calcium-dependent protein binding / calcium ion transport / sperm midpiece / myelin sheath / growth cone / vesicle / protein homotetramerization / transmembrane transporter binding / calmodulin binding / apical plasma membrane / protein domain specific binding / calcium ion binding / centrosome / protein kinase binding / protein-containing complex / mitochondrion / nucleoplasm / metal ion binding / identical protein binding / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |   | |||||||||
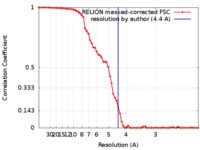
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Hughes TET / Pumroy RA | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Structural insights on TRPV5 gating by endogenous modulators. Authors: Taylor E T Hughes / Ruth A Pumroy / Aysenur Torun Yazici / Marina A Kasimova / Edwin C Fluck / Kevin W Huynh / Amrita Samanta / Sudheer K Molugu / Z Hong Zhou / Vincenzo Carnevale / Tibor ...Authors: Taylor E T Hughes / Ruth A Pumroy / Aysenur Torun Yazici / Marina A Kasimova / Edwin C Fluck / Kevin W Huynh / Amrita Samanta / Sudheer K Molugu / Z Hong Zhou / Vincenzo Carnevale / Tibor Rohacs / Vera Y Moiseenkova-Bell /  Abstract: TRPV5 is a transient receptor potential channel involved in calcium reabsorption. Here we investigate the interaction of two endogenous modulators with TRPV5. Both phosphatidylinositol 4,5- ...TRPV5 is a transient receptor potential channel involved in calcium reabsorption. Here we investigate the interaction of two endogenous modulators with TRPV5. Both phosphatidylinositol 4,5-bisphosphate (PI(4,5)P) and calmodulin (CaM) have been shown to directly bind to TRPV5 and activate or inactivate the channel, respectively. Using cryo-electron microscopy (cryo-EM), we determined TRPV5 structures in the presence of dioctanoyl PI(4,5)P and CaM. The PI(4,5)P structure reveals a binding site between the N-linker, S4-S5 linker and S6 helix of TRPV5. These interactions with PI(4,5)P induce conformational rearrangements in the lower gate, opening the channel. The CaM structure reveals two TRPV5 C-terminal peptides anchoring a single CaM molecule and that calcium inhibition is mediated through a cation-π interaction between Lys116 on the C-lobe of calcium-activated CaM and Trp583 at the intracellular gate of TRPV5. Overall, this investigation provides insight into the endogenous modulation of TRPV5, which has the potential to guide drug discovery. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7967.map.gz emd_7967.map.gz | 25.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7967-v30.xml emd-7967-v30.xml emd-7967.xml emd-7967.xml | 12.8 KB 12.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7967_fsc.xml emd_7967_fsc.xml | 6.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_7967.png emd_7967.png | 80.5 KB | ||
| Filedesc metadata |  emd-7967.cif.gz emd-7967.cif.gz | 6.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7967 http://ftp.pdbj.org/pub/emdb/structures/EMD-7967 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7967 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7967 | HTTPS FTP |
-Related structure data
| Related structure data |  6dmwMC  7965C  7966C  6dmrC  6dmuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7967.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7967.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Calmodulin-bound rbTRPV5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Calmodulin-bound rbTRPV5 tetramer
| Entire | Name: Calmodulin-bound rbTRPV5 tetramer |
|---|---|
| Components |
|
-Supramolecule #1: Calmodulin-bound rbTRPV5 tetramer
| Supramolecule | Name: Calmodulin-bound rbTRPV5 tetramer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|
-Supramolecule #2: rbTRPV5 tetramer
| Supramolecule | Name: rbTRPV5 tetramer / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: rat Calmodulin
| Supramolecule | Name: rat Calmodulin / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Transient receptor potential cation channel subfamily V member 5
| Macromolecule | Name: Transient receptor potential cation channel subfamily V member 5 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 82.899656 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGACPPKAKG PWAQLQKLLI SWPVGEQDWE QYRDRVNMLQ QERIRDSPLL QAAKENDLRL LKILLLNQSC DFQQRGAVGE TALHVAALY DNLEAATLLM EAAPELAKEP ALCEPFVGQT ALHIAVMNQN LNLVRALLAR GASVSARATG AAFRRSPHNL I YYGEHPLS ...String: MGACPPKAKG PWAQLQKLLI SWPVGEQDWE QYRDRVNMLQ QERIRDSPLL QAAKENDLRL LKILLLNQSC DFQQRGAVGE TALHVAALY DNLEAATLLM EAAPELAKEP ALCEPFVGQT ALHIAVMNQN LNLVRALLAR GASVSARATG AAFRRSPHNL I YYGEHPLS FAACVGSEEI VRLLIEHGAD IRAQDSLGNT VLHILILQPN KTFACQMYNL LLSYDEHSDH LQSLELVPNH QG LTPFKLA GVEGNTVMFQ HLMQKRKHVQ WTCGPLTSTL YDLTEIDSWG EELSFLELVV SSKKREARQI LEQTPVKELV SFK WKKYGR PYFCVLASLY ILYMICFTTC CIYRPLKLRD DNRTDPRDIT ILQQKLLQEA YVTHQDNIRL VGELVTVTGA VIIL LLEIP DIFRVGASRY FGQTILGGPF HVIIITYASL VLLTMVMRLT NMNGEVVPLS FALVLGWCSV MYFARGFQML GPFTI MIQK MIFGDLMRFC WLMAVVILGF ASAFHITFQT EDPNNLGEFS DYPTALFSTF ELFLTIIDGP ANYSVDLPFM YCITYA AFA IIATLLMLNL FIAMMGDTHW RVAQERDELW RAQVVATTVM LERKMPRFLW PRSGICGYEY GLGDRWFLRV ENHHDQN PL RVLRYVEAFK CSDKEDGQEQ LSEKRPSTVE SGMLSRASVA FQTPSLSRTT SQSSNSHRGW EILRRNTLGH LNLGLDLG E GDGEEVYHF UniProtKB: Transient receptor potential cation channel subfamily V member 5 |
-Macromolecule #2: Calmodulin-1
| Macromolecule | Name: Calmodulin-1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.852545 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MADQLTEEQI AEFKEAFSLF DKDGDGTITT KELGTVMRSL GQNPTEAELQ DMINEVDADG NGTIDFPEFL TMMARKMKDT DSEEEIREA FRVFDKDGNG YISAAELRHV MTNLGEKLTD EEVDEMIREA DIDGDGQVNY EEFVQMMTAK UniProtKB: Calmodulin-1 |
-Macromolecule #3: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 3 / Number of copies: 3 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8.8 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)