[English] 日本語
 Yorodumi
Yorodumi- EMDB-7870: Structure of the 50S ribosomal subunit from Methicillin Resistant... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7870 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the 50S ribosomal subunit from Methicillin Resistant Staphylococcus aureus in complex with the oxazolidinone antibiotic LZD-6 | |||||||||
 Map data Map data | 50S ribosomal subunit from MRSA in complex with an oxazolidinone antibiotic: LZD-6 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | antibiotic complex / linezolid / oxazolidinone / 50S / ribosome / Ribosome-Antibiotic complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationlarge ribosomal subunit / transferase activity / 5S rRNA binding / ribosomal large subunit assembly / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / negative regulation of translation / rRNA binding ...large ribosomal subunit / transferase activity / 5S rRNA binding / ribosomal large subunit assembly / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / mRNA binding / RNA binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Belousoff MJ / Venugopal H | |||||||||
| Funding support |  Australia, 1 items Australia, 1 items
| |||||||||
 Citation Citation |  Journal: ChemMedChem / Year: 2019 Journal: ChemMedChem / Year: 2019Title: cryoEM-Guided Development of Antibiotics for Drug-Resistant Bacteria. Authors: Matthew J Belousoff / Hari Venugopal / Alexander Wright / Samuel Seoner / Isabella Stuart / Chris Stubenrauch / Rebecca S Bamert / David W Lupton / Trevor Lithgow /  Abstract: While the ribosome is a common target for antibiotics, challenges with crystallography can impede the development of new bioactives using structure-based drug design approaches. In this study we ...While the ribosome is a common target for antibiotics, challenges with crystallography can impede the development of new bioactives using structure-based drug design approaches. In this study we exploit common structural features present in linezolid-resistant forms of both methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus (VRE) to redesign the antibiotic. Enabled by rapid and facile cryoEM structures, this process has identified (S)-2,2-dichloro-N-((3-(3-fluoro-4-morpholinophenyl)-2-oxooxazolidin-5-yl)methyl)acetamide (LZD-5) and (S)-2-chloro-N-((3-(3-fluoro-4-morpholinophenyl)-2-oxooxazolidin-5-yl)methyl) acetamide (LZD-6), which inhibit the ribosomal function and growth of linezolid-resistant MRSA and VRE. The strategy discussed highlights the potential for cryoEM to facilitate the development of novel bioactive materials. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7870.map.gz emd_7870.map.gz | 16.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7870-v30.xml emd-7870-v30.xml emd-7870.xml emd-7870.xml | 39.1 KB 39.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7870.png emd_7870.png | 101.2 KB | ||
| Filedesc metadata |  emd-7870.cif.gz emd-7870.cif.gz | 9.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7870 http://ftp.pdbj.org/pub/emdb/structures/EMD-7870 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7870 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7870 | HTTPS FTP |
-Related structure data
| Related structure data |  6ddgMC  7867C  6dddC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7870.map.gz / Format: CCP4 / Size: 184 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7870.map.gz / Format: CCP4 / Size: 184 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 50S ribosomal subunit from MRSA in complex with an oxazolidinone antibiotic: LZD-6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : 50S Ribosomal subunit from MRSA in complex with oxazolidinone LZD-5
+Supramolecule #1: 50S Ribosomal subunit from MRSA in complex with oxazolidinone LZD-5
+Macromolecule #1: 50S ribosomal protein L19
+Macromolecule #2: 50S ribosomal protein L2
+Macromolecule #3: 50S ribosomal protein L20
+Macromolecule #4: 50S ribosomal protein L21
+Macromolecule #5: 50S ribosomal protein L22
+Macromolecule #6: 50S ribosomal protein L23
+Macromolecule #7: 50S ribosomal protein L24
+Macromolecule #8: 50S ribosomal protein L25
+Macromolecule #9: 50S ribosomal protein L27
+Macromolecule #10: 50S ribosomal protein L28
+Macromolecule #11: 50S ribosomal protein L29
+Macromolecule #12: 50S ribosomal protein L3
+Macromolecule #13: 50S ribosomal protein L30
+Macromolecule #14: 50S ribosomal protein L32
+Macromolecule #15: 50S ribosomal protein L33
+Macromolecule #16: 50S ribosomal protein L34
+Macromolecule #17: 50S ribosomal protein L35
+Macromolecule #18: 50S ribosomal protein L36
+Macromolecule #19: 50S ribosomal protein L4
+Macromolecule #20: 50S ribosomal protein L13
+Macromolecule #21: 50S ribosomal protein L14
+Macromolecule #22: 50S ribosomal protein L15
+Macromolecule #23: 50S ribosomal protein L16
+Macromolecule #24: 50S ribosomal protein L17
+Macromolecule #25: 50S ribosomal protein L18
+Macromolecule #26: 23S rRNA
+Macromolecule #27: 5S rRNA
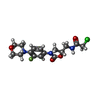
+Macromolecule #28: 2-chloro-N-({(5S)-3-[3-fluoro-4-(morpholin-4-yl)phenyl]-2-oxo-1,3...
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Number classes used: 1 / Applied symmetry - Point group: C1 (asymmetric) / Algorithm: FOURIER SPACE / Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 49223 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)