[English] 日本語
 Yorodumi
Yorodumi- EMDB-7558: MicroED structure of NaK ion channel reveals a process of Na+ par... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7558 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | MicroED structure of NaK ion channel reveals a process of Na+ partition into the selectivity filter | |||||||||
 Map data Map data | NaK ion channel | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ion channel / NaK / TRANSPORT PROTEIN | |||||||||
| Function / homology | Two pore domain potassium channel / Potassium channel domain / Ion channel / potassium channel activity / metal ion binding / identical protein binding / membrane / Transporter / Potassium channel protein Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | electron crystallography / cryo EM / Resolution: 2.5002 Å | |||||||||
 Authors Authors | Liu S / Gonen T | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2018 Journal: Commun Biol / Year: 2018Title: MicroED structure of the NaK ion channel reveals a Na partition process into the selectivity filter. Authors: Shian Liu / Tamir Gonen /  Abstract: Sodium (Na) is a ubiquitous and important inorganic salt mediating many critical biological processes such as neuronal excitation, signaling, and facilitation of various transporters. The hydration ...Sodium (Na) is a ubiquitous and important inorganic salt mediating many critical biological processes such as neuronal excitation, signaling, and facilitation of various transporters. The hydration states of Na are proposed to play critical roles in determining the conductance and the selectivity of Na channels, yet they are rarely captured by conventional structural biology means. Here we use the emerging cryo-electron microscopy (cryoEM) method micro-electron diffraction (MicroED) to study the structure of a prototypical tetrameric Na-conducting channel, NaK, to 2.5 Å resolution from nano-crystals. Two new conformations at the external site of NaK are identified, allowing us to visualize a partially hydrated Na ion at the entrance of the channel pore. A process of dilation coupled with Na movement is identified leading to valuable insights into the mechanism of ion conduction and gating. This study lays the ground work for future studies using MicroED in membrane protein biophysics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7558.map.gz emd_7558.map.gz | 10.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7558-v30.xml emd-7558-v30.xml emd-7558.xml emd-7558.xml | 10.7 KB 10.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7558.png emd_7558.png | 62.9 KB | ||
| Filedesc metadata |  emd-7558.cif.gz emd-7558.cif.gz | 5.1 KB | ||
| Filedesc structureFactors |  emd_7558_sf.cif.gz emd_7558_sf.cif.gz | 509.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7558 http://ftp.pdbj.org/pub/emdb/structures/EMD-7558 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7558 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7558 | HTTPS FTP |
-Validation report
| Summary document |  emd_7558_validation.pdf.gz emd_7558_validation.pdf.gz | 522 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7558_full_validation.pdf.gz emd_7558_full_validation.pdf.gz | 521.6 KB | Display | |
| Data in XML |  emd_7558_validation.xml.gz emd_7558_validation.xml.gz | 4.2 KB | Display | |
| Data in CIF |  emd_7558_validation.cif.gz emd_7558_validation.cif.gz | 4.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7558 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7558 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7558 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7558 | HTTPS FTP |
-Related structure data
| Related structure data |  6cpvMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7558.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7558.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | NaK ion channel | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X: 0.51569 Å / Y: 0.51569 Å / Z: 0.53155 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
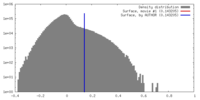
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 79 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : NaK
| Entire | Name: NaK |
|---|---|
| Components |
|
-Supramolecule #1: NaK
| Supramolecule | Name: NaK / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Potassium channel protein
| Macromolecule | Name: Potassium channel protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.706538 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: WKDKEFQVLF VLTILTLISG TIFYSTVEGL RPIDALYFSV VTLTTVGDGN FSPQTDFGKI FTILYIFIGI GLVFGFIHKL AVNVQLPSI LSNLVPR UniProtKB: Transporter |
-Macromolecule #2: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 2 / Number of copies: 12 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #3: (4S)-2-METHYL-2,4-PENTANEDIOL
| Macromolecule | Name: (4S)-2-METHYL-2,4-PENTANEDIOL / type: ligand / ID: 3 / Number of copies: 1 / Formula: MPD |
|---|---|
| Molecular weight | Theoretical: 118.174 Da |
| Chemical component information |  ChemComp-MPD: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 4 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron crystallography |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Average electron dose: 0.1 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Camera length: 1750 mm |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.5002 Å / Resolution method: DIFFRACTION PATTERN/LAYERLINES |
|---|---|
| Crystallography statistics | Number intensities measured: 27479 / Number structure factors: 5643 / Fourier space coverage: 81.7 / R sym: 0.206 / R merge: 0.206 / Overall phase error: 20.27 / Overall phase residual: 20.27 / Phase error rejection criteria: 0 / High resolution: 2.5002 Å / Shell - Shell ID: 1 / Shell - High resolution: 2.5002 Å / Shell - Low resolution: 3.1486 Å / Shell - Number structure factors: 2685 / Shell - Phase residual: 22.68 / Shell - Fourier space coverage: 0.76 / Shell - Multiplicity: 4.1 |
-Atomic model buiding 1
| Refinement | Overall B value: 41 |
|---|---|
| Output model |  PDB-6cpv: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)