[English] 日本語
 Yorodumi
Yorodumi- EMDB-7446: Structure of the DASH/Dam1 complex shows its role at the yeast ki... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7446 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the DASH/Dam1 complex shows its role at the yeast kinetochore-microtubule interface (symmetric reconstruction) | |||||||||
 Map data Map data | DASH/Dam1c symmetric reconstruction | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) | |||||||||
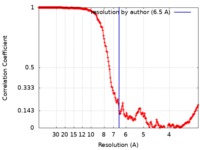
| Method | single particle reconstruction / cryo EM / Resolution: 6.5 Å | |||||||||
 Authors Authors | Jenni S / Harrison SC | |||||||||
 Citation Citation |  Journal: Science / Year: 2018 Journal: Science / Year: 2018Title: Structure of the DASH/Dam1 complex shows its role at the yeast kinetochore-microtubule interface. Authors: Simon Jenni / Stephen C Harrison /  Abstract: Kinetochores connect mitotic-spindle microtubules with chromosomes, allowing microtubule depolymerization to pull chromosomes apart during anaphase while resisting detachment as the microtubule ...Kinetochores connect mitotic-spindle microtubules with chromosomes, allowing microtubule depolymerization to pull chromosomes apart during anaphase while resisting detachment as the microtubule shortens. The heterodecameric DASH/Dam1 complex (DASH/Dam1c), an essential component of yeast kinetochores, assembles into a microtubule-encircling ring. The ring associates with rodlike Ndc80 complexes to organize the kinetochore-microtubule interface. We report the cryo-electron microscopy structure (at ~4.5-angstrom resolution) of a DASH/Dam1c ring and a molecular model of its ordered components, validated by evolutionary direct-coupling analysis. Integrating this structure with that of the Ndc80 complex and with published interaction data yields a molecular picture of kinetochore-microtubule attachment, including how flexible, C-terminal extensions of DASH/Dam1c subunits project and contact widely separated sites on the Ndc80 complex rod and how phosphorylation at previously identified sites might regulate kinetochore assembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7446.map.gz emd_7446.map.gz | 464.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7446-v30.xml emd-7446-v30.xml emd-7446.xml emd-7446.xml | 24 KB 24 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7446_fsc.xml emd_7446_fsc.xml | 21 KB | Display |  FSC data file FSC data file |
| Images |  emd_7446.png emd_7446.png | 156.8 KB | ||
| Masks |  emd_7446_msk_1.map emd_7446_msk_1.map | 512 MB |  Mask map Mask map | |
| Others |  emd_7446_half_map_1.map.gz emd_7446_half_map_1.map.gz emd_7446_half_map_2.map.gz emd_7446_half_map_2.map.gz | 154.4 MB 154.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7446 http://ftp.pdbj.org/pub/emdb/structures/EMD-7446 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7446 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7446 | HTTPS FTP |
-Validation report
| Summary document |  emd_7446_validation.pdf.gz emd_7446_validation.pdf.gz | 78.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7446_full_validation.pdf.gz emd_7446_full_validation.pdf.gz | 77.5 KB | Display | |
| Data in XML |  emd_7446_validation.xml.gz emd_7446_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7446 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7446 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7446 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7446 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7446.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7446.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DASH/Dam1c symmetric reconstruction | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.64 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
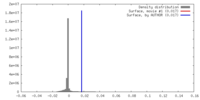
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_7446_msk_1.map emd_7446_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
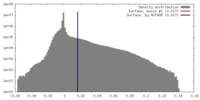
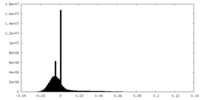
| Density Histograms |
-Half map: Half-map 1 (not masked, not filtered)
| File | emd_7446_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 (not masked, not filtered) | ||||||||||||
| Projections & Slices |
| ||||||||||||
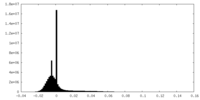
| Density Histograms |
-Half map: Half-map 2 (not masked, not filtered)
| File | emd_7446_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 (not masked, not filtered) | ||||||||||||
| Projections & Slices |
| ||||||||||||
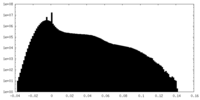
| Density Histograms |
- Sample components
Sample components
+Entire : DASH/Dam1 complex
+Supramolecule #1: DASH/Dam1 complex
+Macromolecule #1: Ask1
+Macromolecule #2: Dad1
+Macromolecule #3: Dad2
+Macromolecule #4: Dad3
+Macromolecule #5: Dad4
+Macromolecule #6: Dam1
+Macromolecule #7: Duo1
+Macromolecule #8: Hsk3
+Macromolecule #9: Spc19
+Macromolecule #10: Spc34
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 2.6 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: C-flat-1.2/1.3 4C / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 89 % / Chamber temperature: 293 K / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Number real images: 2723 / Average electron dose: 1.115 e/Å2 Details: Movies collected: 50 frames with 1.115 e/A2/per frame |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 30488 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)