[English] 日本語
 Yorodumi
Yorodumi- EMDB-71708: Alpha1/Beta Heteromeric Glycine receptor in the presence of 0.200... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Alpha1/Beta Heteromeric Glycine receptor in the presence of 0.200 mM strychnine and 0.02 mM ivermectin | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ion Channel / Pentameric / Glycine / Receptor / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationextracellularly glycine-gated ion channel activity / extracellularly glycine-gated chloride channel activity / transmitter-gated monoatomic ion channel activity / regulation of neuron differentiation / glycine binding / ligand-gated monoatomic ion channel activity / response to amino acid / chloride channel complex / monoatomic ion transport / central nervous system development ...extracellularly glycine-gated ion channel activity / extracellularly glycine-gated chloride channel activity / transmitter-gated monoatomic ion channel activity / regulation of neuron differentiation / glycine binding / ligand-gated monoatomic ion channel activity / response to amino acid / chloride channel complex / monoatomic ion transport / central nervous system development / transmembrane signaling receptor activity / intracellular protein localization / perikaryon / postsynaptic membrane / dendrite / metal ion binding / membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.89 Å | |||||||||
 Authors Authors | Gibbs E / Chakrapani S | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Alpha1/Beta Heteromeric Glycine receptor in the presence of 0.200 mM strychnine and 0.02 mM ivermectin Authors: Gibbs E / Chakrapani S | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_71708.map.gz emd_71708.map.gz | 95.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-71708-v30.xml emd-71708-v30.xml emd-71708.xml emd-71708.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_71708_fsc.xml emd_71708_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_71708.png emd_71708.png | 90.9 KB | ||
| Masks |  emd_71708_msk_1.map emd_71708_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-71708.cif.gz emd-71708.cif.gz | 6.4 KB | ||
| Others |  emd_71708_half_map_1.map.gz emd_71708_half_map_1.map.gz emd_71708_half_map_2.map.gz emd_71708_half_map_2.map.gz | 80.8 MB 80.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-71708 http://ftp.pdbj.org/pub/emdb/structures/EMD-71708 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-71708 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-71708 | HTTPS FTP |
-Related structure data
| Related structure data |  9pkzMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_71708.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_71708.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_71708_msk_1.map emd_71708_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_71708_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_71708_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Glycine receptor alpha1/beta
| Entire | Name: Glycine receptor alpha1/beta |
|---|---|
| Components |
|
-Supramolecule #1: Glycine receptor alpha1/beta
| Supramolecule | Name: Glycine receptor alpha1/beta / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Glycine receptor, alpha 1
| Macromolecule | Name: Glycine receptor, alpha 1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 52.537598 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MFALGIYLWE TIVFFSLAAS QQAAARKAAS PMPPSEFLDK LMGKVSGYDA RIRPNFKGPP VNVTCNIFIN SFGSIAETTM DYRVNIFLR QQWNDPRLAY SEYPDDSLDL DPSMLDSIWK PDLFFANEKG ANFHEVTTDN KLLRISKNGN VLYSIRITLV L ACPMDLKN ...String: MFALGIYLWE TIVFFSLAAS QQAAARKAAS PMPPSEFLDK LMGKVSGYDA RIRPNFKGPP VNVTCNIFIN SFGSIAETTM DYRVNIFLR QQWNDPRLAY SEYPDDSLDL DPSMLDSIWK PDLFFANEKG ANFHEVTTDN KLLRISKNGN VLYSIRITLV L ACPMDLKN FPMDVQTCIM QLESFGYTMN DLIFEWDEKG AVQVADGLTL PQFILKEEKD LRYCTKHYNT GKFTCIEARF HL ERQMGYY LIQMYIPSLL IVILSWVSFW INMDAAPARV GLGITTVLTM TTQSSGSRAS LPKVSYVKAI DIWMAVCLLF VFS ALLEYA AVNFIARQHK ELLRFQRRRR HLKEDEAGDG RFSFAAYGMG PACLQAKDGM AIKGNNNNAP TSTNPPEKTV EEMR KLFIS RAKRIDTVSR VAFPLVFLIF NIFYWITYKI IRSEDIHKQL VPRGSHHHHH HHH UniProtKB: Glycine receptor, alpha 1 |
-Macromolecule #2: Glycine receptor beta subunit 2
| Macromolecule | Name: Glycine receptor beta subunit 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 65.820281 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MKALKVIFML LIICLWMEGG FTKEKSAKKW SHPQFEKGGG SGGGSGGGSW SHPQFEKGGG SGGGSGGGSW SHPQFEKGGG SGGGSGGGS WSHPQFEKEN LYFQGEKSAK KGKKKGKQVY CPSQLSSEDL ARVPANSTSN ILNKLLITYD PRIRPNFKGI P VEDRVNIF ...String: MKALKVIFML LIICLWMEGG FTKEKSAKKW SHPQFEKGGG SGGGSGGGSW SHPQFEKGGG SGGGSGGGSW SHPQFEKGGG SGGGSGGGS WSHPQFEKEN LYFQGEKSAK KGKKKGKQVY CPSQLSSEDL ARVPANSTSN ILNKLLITYD PRIRPNFKGI P VEDRVNIF INSFGSIQET TMDYRVNIFL RQRWNDPRLR LPQDFKSDSL TVDPKMFKCL WKPDLFFANE KSANFHDVTQ EN ILLFIFR NGDVLISMRL SVTLSCPLDL TLFPMDTQRC KMQLESFGYT TDDLQFMWQS GDPVQMDEIA LPQFDIKQED IEY GNCTKY YAGTGYYTCV EVIFTLRRQV GFYMMGVYAP TLLIVVLSWL SFWINPDASA ARVPLGILSV LSLSSECTSL ASEL PKVSY VKAIDIWLIA CLLFGFASLV EYAVVQVMLN SPKLLEAERA KIATKEKAEG KTPAKNTING MGSTPIHVST LQVTE TRCK KVCTSKSDLR TNDFSIVGSL PRDFELSNFD CYGKPIEVGS AFSKSQAKNN KKPPPPKPVI PSAAKRIDLY ARALFP FSF LFFNVIYWSV YLENLYFQGT ETSQVAPA UniProtKB: Glycine receptor subunit beta |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 5 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)...
| Macromolecule | Name: (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H- ...Name: (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside type: ligand / ID: 4 / Number of copies: 5 / Formula: IVM |
|---|---|
| Molecular weight | Theoretical: 875.093 Da |
-Macromolecule #5: STRYCHNINE
| Macromolecule | Name: STRYCHNINE / type: ligand / ID: 5 / Number of copies: 5 / Formula: SY9 |
|---|---|
| Molecular weight | Theoretical: 334.412 Da |
| Chemical component information | 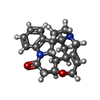 ChemComp-SY9: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































