+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7129 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human PI4KIIIa lipid kinase complex | |||||||||
 Map data Map data | Human PI4KIIIa lipid kinase complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | kinase / phosphoinositide synthesis / Transferase-Signaling Protein complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationreorganization of cellular membranes to establish viral sites of replication / Synthesis of PIPs at the ER membrane / 1-phosphatidylinositol 4-kinase / 1-phosphatidylinositol 4-kinase activity / Schwann cell migration / Synthesis of PIPs at the Golgi membrane / modulation by host of viral process / Golgi-associated vesicle membrane / myelination in peripheral nervous system / phosphatidylinositol biosynthetic process ...reorganization of cellular membranes to establish viral sites of replication / Synthesis of PIPs at the ER membrane / 1-phosphatidylinositol 4-kinase / 1-phosphatidylinositol 4-kinase activity / Schwann cell migration / Synthesis of PIPs at the Golgi membrane / modulation by host of viral process / Golgi-associated vesicle membrane / myelination in peripheral nervous system / phosphatidylinositol biosynthetic process / phosphatidylinositol-mediated signaling / phosphatidylinositol phosphate biosynthetic process / myelination / protein localization to plasma membrane / actin cytoskeleton organization / neuron projection / cadherin binding / focal adhesion / signal transduction / extracellular exosome / ATP binding / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
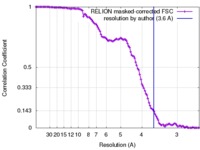
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Lees JA / Zhang Y | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2017 Journal: Proc Natl Acad Sci U S A / Year: 2017Title: Architecture of the human PI4KIIIα lipid kinase complex. Authors: Joshua A Lees / Yixiao Zhang / Michael S Oh / Curtis M Schauder / Xiaoling Yu / Jeremy M Baskin / Kerry Dobbs / Luigi D Notarangelo / Pietro De Camilli / Thomas Walz / Karin M Reinisch /  Abstract: Plasma membrane (PM) phosphoinositides play essential roles in cell physiology, serving as both markers of membrane identity and signaling molecules central to the cell's interaction with its ...Plasma membrane (PM) phosphoinositides play essential roles in cell physiology, serving as both markers of membrane identity and signaling molecules central to the cell's interaction with its environment. The first step in PM phosphoinositide synthesis is the conversion of phosphatidylinositol (PI) to PI4P, the precursor of PI(4,5)P and PI(3,4,5)P This conversion is catalyzed by the PI4KIIIα complex, comprising a lipid kinase, PI4KIIIα, and two regulatory subunits, TTC7 and FAM126. We here report the structure of this complex at 3.6-Å resolution, determined by cryo-electron microscopy. The proteins form an obligate ∼700-kDa superassembly with a broad surface suitable for membrane interaction, toward which the kinase active sites are oriented. The structural complexity of the assembly highlights PI4P synthesis as a major regulatory junction in PM phosphoinositide homeostasis. Our studies provide a framework for further exploring the mechanisms underlying PM phosphoinositide regulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7129.map.gz emd_7129.map.gz | 9.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7129-v30.xml emd-7129-v30.xml emd-7129.xml emd-7129.xml | 24.9 KB 24.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7129_fsc.xml emd_7129_fsc.xml | 11.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_7129.png emd_7129.png | 68 KB | ||
| Filedesc metadata |  emd-7129.cif.gz emd-7129.cif.gz | 7.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7129 http://ftp.pdbj.org/pub/emdb/structures/EMD-7129 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7129 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7129 | HTTPS FTP |
-Related structure data
| Related structure data |  6bq1MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7129.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7129.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human PI4KIIIa lipid kinase complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
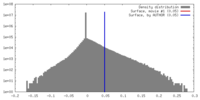
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human phosphatidylinositol 4-kinase III alpha complex
| Entire | Name: Human phosphatidylinositol 4-kinase III alpha complex |
|---|---|
| Components |
|
-Supramolecule #1: Human phosphatidylinositol 4-kinase III alpha complex
| Supramolecule | Name: Human phosphatidylinositol 4-kinase III alpha complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 700 KDa |
-Macromolecule #1: Phosphatidylinositol 4-kinase III alpha (PI4KA)
| Macromolecule | Name: Phosphatidylinositol 4-kinase III alpha (PI4KA) / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: 1-phosphatidylinositol 4-kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 173.821438 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)MMQCV IAVADKVFDA FLNMM ADKA KTKENEEELE RHAQFLLVNF NHIHKRIRRV ADKYLSGLVD KFPHLLWSGT VLKTMLDILQ TLSLSLSADI HKDQPY YDI PDAPYRITVP DTYEARESIV KDFAARCGMI LQEAMKWAPT VTKSHLQEYL NKHQNWVSGL SQHTGLAMAT ESILHFA GY NKQNTTLGAT QLSERPACVK KDYSNFMASL NLRNRYAGEV YGMIRFSGTT GQMSDLNKMM VQDLHSALDR SHPQHYTQ A MFKLTAMLIS SKDCDPQLLH HLCWGPLRMF NEHGMETALA CWEWLLAGKD GVEVPFMREM AGAWHMTVEQ KFGLFSAEI KEADPLAASE ASQPKPCPPE VTPHYIWIDF LVQRFEIAKY CSSDQVEIFS SLLQRSMSLN IGGAKGSMNR HVAAIGPRFK LLTLGLSLL HADVVPNATI RNVLREKIYS TAFDYFSCPP KFPTQGEKRL REDISIMIKF WTAMFSDKKY LTASQLVPPD N QDTRSNLD ITVGSRQQAT QGWINTYPLS SGMSTISKKS GMSKKTNRGS QLHKYYMKRR TLLLSLLATE IERLITWYNP LS APELELD QAGENSVANW RSKYISLSEK QWKDNVNLAW SISPYLAVQL PARFKNTEAI GNEVTRLVRL DPGAVSDVPE AIK FLVTWH TIDADAPELS HVLCWAPTDP PTGLSYFSSM YPPHPLTAQY GVKVLRSFPP DAILFYIPQI VQALRYDKMG YVRE YILWA ASKSQLLAHQ FIWNMKTNIY LDEEGHQKDP DIGDLLDQLV EEITGSLSGP AKDFYQREFD FFNKITNVSA IIKPY PKGD ERKKACLSAL SEVKVQPGCY LPSNPEAIVL DIDYKSGTPM QSAAKAPYLA KFKVKRCGVS ELEKEGLRCR SDSEDE CST QEADGQKISW QAAIFKVGDD CRQDMLALQI IDLFKNIFQL VGLDLFVFPY RVVATAPGCG VIECIPDCTS RDQLGRQ TD FGMYDYFTRQ YGDESTLAFQ QARYNFIRSM AAYSLLLFLL QIKDRHNGNI MLDKKGHIIH IDFGFMFESS PGGNLGWE P DIKLTDEMVM IMGGKMEATP FKWFMEMCVR GYLAVRPYMD AVVSLVTLML DTGLPCFRGQ TIKLLKHRFS PNMTEREAA NFIMKVIQSC FLSNRSRTYD MIQYYQNDIP Y UniProtKB: Phosphatidylinositol 4-kinase alpha |
-Macromolecule #2: Tetratricopeptide repeat protein 7B
| Macromolecule | Name: Tetratricopeptide repeat protein 7B / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 96.46057 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MYPYDVPDYA AIAGAPDLKL MATKKAGSRL ETEIERCRSE CQWERIPELV KQLSAKLIAN DDMAELLLGE SKLEQYLKEH PLRQGASPR GPKPQLTEVR KHLTAALDRG NLKSEFLQES NLIMAKLNYV EGDYKEALNI YARVGLDDLP LTAVPPYRLR V IAEAYATK ...String: MYPYDVPDYA AIAGAPDLKL MATKKAGSRL ETEIERCRSE CQWERIPELV KQLSAKLIAN DDMAELLLGE SKLEQYLKEH PLRQGASPR GPKPQLTEVR KHLTAALDRG NLKSEFLQES NLIMAKLNYV EGDYKEALNI YARVGLDDLP LTAVPPYRLR V IAEAYATK GLCLEKLPIS SSTSNLHVDR EQDVITCYEK AGDIALLYLQ EIERVILSNI QNRSPKPGPA PHDQELGFFL ET GLQRAHV LYFKNGNLTR GVGRFRELLR AVETRTTQNL RMTIARQLAE ILLRGMCEQS YWNPLEDPPC QSPLDDPLRK GAN TKTYTL TRRARVYSGE NIFCPQENTE EALLLLLISE SMANRDAVLS RIPEHKSDRL ISLQSASVVY DLLTIALGRR GQYE MLSEC LERAMKFAFE EFHLWYQFAL SLMAAGKSAR AVKVLKECIR LKPDDATIPL LAAKLCMGSL HWLEEAEKFA KTVVD VGEK TSEFKAKGYL ALGLTYSLQA TDASLRGMQE VLQRKALLAF QRAHSLSPTD HQAAFYLALQ LAISRQIPEA LGYVRQ ALQ LQGDDANSLH LLALLLSAQK HYHDALNIID MALSEYPENF ILLFSKVKLQ SLCRGPDEAL LTCKHMLQIW KSCYNLT NP SDSGRGSSLL DRTIADRRQL NTITLPDFSD PETGSVHATS VAASRVEQAL SEVASSLQSS APKQGPLHPW MTLAQIWL H AAEVYIGIGK PAEATACTQE AANLFPMSHN VLYMRGQIAE LRGSMDEARR WYEEALAISP THVKSMQRLA LILHQLGRY SLAEKILRDA VQVNSTAHEV WNGLGEVLQA QGNDAAATEC FLTALELEAS SPAVPFTIIP RVL UniProtKB: Tetratricopeptide repeat protein 7B |
-Macromolecule #3: Protein FAM126A
| Macromolecule | Name: Protein FAM126A / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 32.688514 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GPGSFTSEKG VVEEWLSEFK TLPETSLPNY ATNLKDKSSL VSSLYKVIQE PQSELLEPVC HQLFEFYRSG EEQLLQFTLQ FLPELIWCY LAVSASRNVH SSGCIEALLL GVYNLEIVDK QGHTKVLSFT IPSLSKPSVY HEPSSIGSMA LTESALSQHG L SKVVYSGP ...String: GPGSFTSEKG VVEEWLSEFK TLPETSLPNY ATNLKDKSSL VSSLYKVIQE PQSELLEPVC HQLFEFYRSG EEQLLQFTLQ FLPELIWCY LAVSASRNVH SSGCIEALLL GVYNLEIVDK QGHTKVLSFT IPSLSKPSVY HEPSSIGSMA LTESALSQHG L SKVVYSGP HPQREMLTAQ NRFEVLTFLL LCYNAALTYM PSVSLQSLCQ ICSRICVCGY PRQHVRKYKG ISSRIPVSSG FM VQMLTGI YFAFYNGEWD LAQKALDDII YRAQLELYPE PLLVANAIKA SLPHG UniProtKB: Hyccin |
-Macromolecule #4: Phosphatidylinositol 4-kinase III alpha (PI4KA)
| Macromolecule | Name: Phosphatidylinositol 4-kinase III alpha (PI4KA) / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO / EC number: 1-phosphatidylinositol 4-kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 173.906547 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)MMQC VIAVADKVFD A FLNMMADK AKTKENEEEL ERHAQFLLVN FNHIHKRIRR VADKYLSGLV DKFPHLLWSG TVLKTMLDIL QTLSLSLSAD IH KDQPYYD IPDAPYRITV PDTYEARESI VKDFAARCGM ILQEAMKWAP TVTKSHLQEY LNKHQNWVSG LSQHTGLAMA TES ILHFAG YNKQNTTLGA TQLSERPACV KKDYSNFMAS LNLRNRYAGE VYGMIRFSGT TGQMSDLNKM MVQDLHSALD RSHP QHYTQ AMFKLTAMLI SSKDCDPQLL HHLCWGPLRM FNEHGMETAL ACWEWLLAGK DGVEVPFMRE MAGAWHMTVE QKFGL FSAE IKEADPLAAS EASQPKPCPP EVTPHYIWID FLVQRFEIAK YCSSDQVEIF SSLLQRSMSL NIGGAKGSMN RHVAAI GPR FKLLTLGLSL LHADVVPNAT IRNVLREKIY STAFDYFSCP PKFPTQGEKR LREDISIMIK FWTAMFSDKK YLTASQL VP PDNQDTRSNL DITVGSRQQA TQGWINTYPL SSGMSTISKK SGMSKKTNRG SQLHKYYMKR RTLLLSLLAT EIERLITW Y NPLSAPELEL DQAGENSVAN WRSKYISLSE KQWKDNVNLA WSISPYLAVQ LPARFKNTEA IGNEVTRLVR LDPGAVSDV PEAIKFLVTW HTIDADAPEL SHVLCWAPTD PPTGLSYFSS MYPPHPLTAQ YGVKVLRSFP PDAILFYIPQ IVQALRYDKM GYVREYILW AASKSQLLAH QFIWNMKTNI YLDEEGHQKD PDIGDLLDQL VEEITGSLSG PAKDFYQREF DFFNKITNVS A IIKPYPKG DERKKACLSA LSEVKVQPGC YLPSNPEAIV LDIDYKSGTP MQSAAKAPYL AKFKVKRCGV SELEKEGLRC RS DSEDECS TQEADGQKIS WQAAIFKVGD DCRQDMLALQ IIDLFKNIFQ LVGLDLFVFP YRVVATAPGC GVIECIPDCT SRD QLGRQT DFGMYDYFTR QYGDESTLAF QQARYNFIRS MAAYSLLLFL LQIKDRHNGN IMLDKKGHII HIDFGFMFES SPGG NLGWE PDIKLTDEMV MIMGGKMEAT PFKWFMEMCV RGYLAVRPYM DAVVSLVTLM LDTGLPCFRG QTIKLLKHRF SPNMT EREA ANFIMKVIQS CFLSNRSRTY DMIQYYQNDI PY UniProtKB: Phosphatidylinositol 4-kinase alpha |
-Macromolecule #5: 5-{2-amino-1-[4-(morpholin-4-yl)phenyl]-1H-benzimidazol-6-yl}-N-(...
| Macromolecule | Name: 5-{2-amino-1-[4-(morpholin-4-yl)phenyl]-1H-benzimidazol-6-yl}-N-(2-fluorophenyl)-2-methoxypyridine-3-sulfonamide type: ligand / ID: 5 / Number of copies: 2 / Formula: E4S |
|---|---|
| Molecular weight | Theoretical: 574.626 Da |
| Chemical component information | 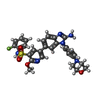 ChemComp-E4S: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.25 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-40 / Number real images: 3280 / Average exposure time: 10.0 sec. / Average electron dose: 47.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 3.7 µm / Calibrated defocus min: 1.8 µm / Calibrated magnification: 38461 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)