+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human MPC matrix-open | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpyruvate import into mitochondria / inner mitochondrial membrane protein complex / pyruvate transmembrane transporter activity / pyruvate decarboxylation to acetyl-CoA / Pyruvate metabolism / detection of maltose stimulus / maltose transport complex / carbohydrate transport / carbohydrate transmembrane transporter activity / maltose binding ...pyruvate import into mitochondria / inner mitochondrial membrane protein complex / pyruvate transmembrane transporter activity / pyruvate decarboxylation to acetyl-CoA / Pyruvate metabolism / detection of maltose stimulus / maltose transport complex / carbohydrate transport / carbohydrate transmembrane transporter activity / maltose binding / maltose transport / maltodextrin transmembrane transport / positive regulation of insulin secretion involved in cellular response to glucose stimulus / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / ATP-binding cassette (ABC) transporter complex / cell chemotaxis / outer membrane-bounded periplasmic space / periplasmic space / mitochondrial inner membrane / DNA damage response / mitochondrion / identical protein binding / nucleus / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.57 Å | |||||||||
 Authors Authors | Qi X / Sun Y / Wang Y | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure of human MPC matrix-open Authors: Qi X / Sun Y / Wang Y | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_70261.map.gz emd_70261.map.gz | 136.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-70261-v30.xml emd-70261-v30.xml emd-70261.xml emd-70261.xml | 11.7 KB 11.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_70261.png emd_70261.png | 90.3 KB | ||
| Filedesc metadata |  emd-70261.cif.gz emd-70261.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-70261 http://ftp.pdbj.org/pub/emdb/structures/EMD-70261 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-70261 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-70261 | HTTPS FTP |
-Validation report
| Summary document |  emd_70261_validation.pdf.gz emd_70261_validation.pdf.gz | 476.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_70261_full_validation.pdf.gz emd_70261_full_validation.pdf.gz | 475.7 KB | Display | |
| Data in XML |  emd_70261_validation.xml.gz emd_70261_validation.xml.gz | 7 KB | Display | |
| Data in CIF |  emd_70261_validation.cif.gz emd_70261_validation.cif.gz | 8.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-70261 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-70261 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-70261 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-70261 | HTTPS FTP |
-Related structure data
| Related structure data |  9o9sMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_70261.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_70261.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.738 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : MPC1-2
| Entire | Name: MPC1-2 |
|---|---|
| Components |
|
-Supramolecule #1: MPC1-2
| Supramolecule | Name: MPC1-2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Mitochondrial pyruvate carrier 1/MBP chimera protein
| Macromolecule | Name: Mitochondrial pyruvate carrier 1/MBP chimera protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.069273 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAGALVRKAA DYVRSKDFRD YLMSTHFWGP VANWGLPIAA INDMKKSPEI ISGRMTFALC CYSLTFMRFA YKVQPRNWLL FACHATNEV AQLIQGGRLI KHEMTKTASA GGGGSGGGGS GGGGSEEGKL VIWINGDKGY NGLAEVGKKF EKDTGIKVTV E HPDKLEEK ...String: MAGALVRKAA DYVRSKDFRD YLMSTHFWGP VANWGLPIAA INDMKKSPEI ISGRMTFALC CYSLTFMRFA YKVQPRNWLL FACHATNEV AQLIQGGRLI KHEMTKTASA GGGGSGGGGS GGGGSEEGKL VIWINGDKGY NGLAEVGKKF EKDTGIKVTV E HPDKLEEK FPQVAATGDG PDIIFWAHDR FGGYAQSGLL AEITPDKAFQ DKLYPFTWDA VRYNGKLIAY PIAVEALSLI YN KDLLPNP PKTWEEIPAL DKELKAKGKS ALMFNLQEPY FTWPLIAADG GYAFKYENGK YDIKDVGVDN AGAKAGLTFL IDL IKNKHM NADTDYSIAE AAFNKGETAM TINGPWAWSN IDTSKVNYGV TVLPTFKGQP SKPFVGVLSA GINAASPNKE LAKE FLENY LLTDEGLEAV NKDKPLGAVA LKSYEEELVK DPRVAATMEN AQKGEIMPNI PQMSAFWYAV RTAVINAASG RQTVD AALA AAQ UniProtKB: Mitochondrial pyruvate carrier 1, Maltose/maltodextrin-binding periplasmic protein |
-Macromolecule #2: Mitochondrial pyruvate carrier 2
| Macromolecule | Name: Mitochondrial pyruvate carrier 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 30.825439 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSAAGARGLR ATYHRLLDKV ELMLPEKLRP LYNHPAGPRT VFFWAPIMKW GLVCAGLADM ARPAEKLSTA QSAVLMATGF IWSRYSLVI IPKNWSLFAV NFFVGAAGAS QLFRIWRYNQ ELKAKDLGRK LLEAARAGQL DEVRILLANG ADVNAADNTG T TPLHLAAY ...String: MSAAGARGLR ATYHRLLDKV ELMLPEKLRP LYNHPAGPRT VFFWAPIMKW GLVCAGLADM ARPAEKLSTA QSAVLMATGF IWSRYSLVI IPKNWSLFAV NFFVGAAGAS QLFRIWRYNQ ELKAKDLGRK LLEAARAGQL DEVRILLANG ADVNAADNTG T TPLHLAAY SGHLEIVEVL LKHGADVDAS DVFGYTPLHL AAYWGHLEIV EVLLKNGADV NAMDSDGMTP LHLAAKWGYL EI VEVLLKH GADVNAQDKF GKTAFDISID NGNEDLAEIL QKLN UniProtKB: Mitochondrial pyruvate carrier 2 |
-Macromolecule #3: (E)-2-cyano-3-(1-phenylindol-3-yl)prop-2-enoic acid
| Macromolecule | Name: (E)-2-cyano-3-(1-phenylindol-3-yl)prop-2-enoic acid / type: ligand / ID: 3 / Number of copies: 1 / Formula: I2R |
|---|---|
| Molecular weight | Theoretical: 288.3 Da |
| Chemical component information | 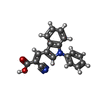 ChemComp-I2R: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)




















