+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6957 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Doublet microtubule of zebrafish sperm axoneme, ktu_null mutant | |||||||||
 Map data Map data | doublet microtubule structure from zebrafish sperm flagella, ktu_null mutant | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
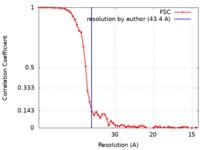
| Method | electron tomography / cryo EM / Resolution: 43.4 Å | |||||||||
 Authors Authors | Yamaguchi H / Oda T / Kikkawa M / Takeda H | |||||||||
 Citation Citation |  Journal: Elife / Year: 2018 Journal: Elife / Year: 2018Title: Systematic studies of all PIH proteins in zebrafish reveal their distinct roles in axonemal dynein assembly. Authors: Hiroshi Yamaguchi / Toshiyuki Oda / Masahide Kikkawa / Hiroyuki Takeda /  Abstract: Construction of motile cilia/flagella requires cytoplasmic preassembly of axonemal dyneins before transport into cilia. Axonemal dyneins have various subtypes, but the roles of each dynein subtype ...Construction of motile cilia/flagella requires cytoplasmic preassembly of axonemal dyneins before transport into cilia. Axonemal dyneins have various subtypes, but the roles of each dynein subtype and their assembly processes remain elusive in vertebrates. The PIH protein family, consisting of four members, has been implicated in the assembly of different dynein subtypes, although evidence for this idea is sparse. Here, we established zebrafish mutants of all four PIH-protein genes: , , , and , and analyzed the structures of axonemal dyneins in mutant spermatozoa by cryo-electron tomography. Mutations caused the loss of specific dynein subtypes, which was correlated with abnormal sperm motility. We also found organ-specific compositions of dynein subtypes, which could explain the severe motility defects of mutant Kupffer's vesicle cilia. Our data demonstrate that all vertebrate PIH proteins are differently required for cilia/flagella motions and the assembly of axonemal dyneins, assigning specific dynein subtypes to each PIH protein. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6957.map.gz emd_6957.map.gz | 9.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6957-v30.xml emd-6957-v30.xml emd-6957.xml emd-6957.xml | 10.6 KB 10.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6957_fsc.xml emd_6957_fsc.xml | 6.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_6957.png emd_6957.png | 100.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6957 http://ftp.pdbj.org/pub/emdb/structures/EMD-6957 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6957 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6957 | HTTPS FTP |
-Validation report
| Summary document |  emd_6957_validation.pdf.gz emd_6957_validation.pdf.gz | 78.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6957_full_validation.pdf.gz emd_6957_full_validation.pdf.gz | 77.8 KB | Display | |
| Data in XML |  emd_6957_validation.xml.gz emd_6957_validation.xml.gz | 495 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6957 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6957 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6957 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6957 | HTTPS FTP |
-Related structure data
| Related structure data |  6954C  6955C  6956C  6958C  6959C  6960C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6957.map.gz / Format: CCP4 / Size: 9.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6957.map.gz / Format: CCP4 / Size: 9.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | doublet microtubule structure from zebrafish sperm flagella, ktu_null mutant | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 7.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
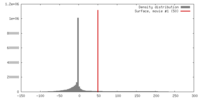
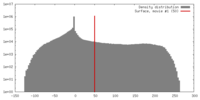
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Doublet microtubule from zebrafish sperm flagella, ktu_null mutant
| Entire | Name: Doublet microtubule from zebrafish sperm flagella, ktu_null mutant |
|---|---|
| Components |
|
-Supramolecule #1: Doublet microtubule from zebrafish sperm flagella, ktu_null mutant
| Supramolecule | Name: Doublet microtubule from zebrafish sperm flagella, ktu_null mutant type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 | ||||||
|---|---|---|---|---|---|---|---|
| Grid | Model: Homemade / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | ||||||
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM GP | ||||||
| Sectioning | Other: NO SECTIONING | ||||||
| Fiducial marker |
|
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3100FFC |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter |
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Average electron dose: 1.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 30000 |
| Sample stage | Specimen holder model: GATAN 914 HIGH TILT LIQUID NITROGEN CRYO TRANSFER TOMOGRAPHY HOLDER Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)