+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6900 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cASIC+Mambalgin1 | |||||||||
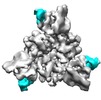
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |   Dendroaspis polylepis polylepis (black mamba) / Gallus gallus Dendroaspis polylepis polylepis (black mamba) / Gallus gallus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.4 Å | |||||||||
 Authors Authors | Sun D / Yu Y / Xue X | |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2018 Journal: Cell Discov / Year: 2018Title: Cryo-EM structure of the ASIC1a-mambalgin-1 complex reveals that the peptide toxin mambalgin-1 inhibits acid-sensing ion channels through an unusual allosteric effect. Authors: Demeng Sun / You Yu / Xiaobin Xue / Man Pan / Ming Wen / Siyu Li / Qian Qu / Xiaorun Li / Longhua Zhang / Xueming Li / Lei Liu / Maojun Yang / Changlin Tian /  Abstract: Acid-sensing ion channels (ASICs) are neuronal voltage-independent Na channels that are activated by extracellular acidification. ASICs play essential roles in a wide range of physiological ...Acid-sensing ion channels (ASICs) are neuronal voltage-independent Na channels that are activated by extracellular acidification. ASICs play essential roles in a wide range of physiological processes, including sodium homeostasis, synaptic plasticity, neurodegeneration, and sensory transduction. Mambalgins, a family of three-finger toxins isolated from black mamba venom, specifically inhibit ASICs to exert strong analgesic effects in vivo, thus are thought to have potential therapeutic values against pain. However, the interaction and inhibition mechanism of mambalgin on ASICs remains elusive. Here, we report a cryo-electron microscopy (cryo-EM) structure of chicken ASIC1a (cASIC1a) in complex with mambalgin-1 toxin at 5.4 Å resolution. Our structure provides the first experimental evidence that mambalgin-1 interacts directly with the extracellular thumb domain of cASIC1a, rather than inserting into the acid-sensing pocket, as previously reported. Binding of mambalgin-1 leads to relocation of the thumb domain that could disrupt the acidic pocket of cASIC1a, illustrating an unusual inhibition mechanism of toxins on ASIC channels through an allosteric effect. These findings establish a structural basis for the toxicity of the mambalgins, and provide crucial insights for the development of new optimized inhibitors of ASICs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6900.map.gz emd_6900.map.gz | 1.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6900-v30.xml emd-6900-v30.xml emd-6900.xml emd-6900.xml | 9.9 KB 9.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6900.png emd_6900.png | 108.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6900 http://ftp.pdbj.org/pub/emdb/structures/EMD-6900 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6900 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6900 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6900.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6900.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.64 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : cASIC and Mambalgin1
| Entire | Name: cASIC and Mambalgin1 |
|---|---|
| Components |
|
-Supramolecule #1: cASIC and Mambalgin1
| Supramolecule | Name: cASIC and Mambalgin1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: cASIC
| Supramolecule | Name: cASIC / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Dendroaspis polylepis polylepis (black mamba) Dendroaspis polylepis polylepis (black mamba) |
-Supramolecule #3: Mambalgin1
| Supramolecule | Name: Mambalgin1 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|
-Macromolecule #1: cASIC
| Macromolecule | Name: cASIC / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: Gallus gallus |
| Sequence | String: YPHVTKLDEV AATRLTFPAV TFCNLNEFRF SRVTKNDLYH AGELLALLNN RYEIPDTQTA DEKQLEILQD KANFRNFKPK PFNMLEFYDR AGHDIREMLL SCFFRGEQCS PEDFKVVFTR YGKCYTFNAG QDGKPRLITM KGGTGNGLEI MLDIQQDEYL PVWGETDETS ...String: YPHVTKLDEV AATRLTFPAV TFCNLNEFRF SRVTKNDLYH AGELLALLNN RYEIPDTQTA DEKQLEILQD KANFRNFKPK PFNMLEFYDR AGHDIREMLL SCFFRGEQCS PEDFKVVFTR YGKCYTFNAG QDGKPRLITM KGGTGNGLEI MLDIQQDEYL PVWGETDETS FEAGIKVQIH SQDEPPLIDQ LGFGVAPGFQ TFVSCQEQRL IYLPPPWGDC KATTGDDTYS ITACRIDCET RYLVENCNCR MVHMPGDAPY CTPEQYKECA DPALDFLVEK DNEYCVCEMP CNVTRYGKEL SMVKIPSKAS AKYLAKKYNK SEQYIGENIL VLDIFFEALN YETIEQKK |
-Macromolecule #2: Mambalgin1
| Macromolecule | Name: Mambalgin1 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Dendroaspis polylepis polylepis (black mamba) Dendroaspis polylepis polylepis (black mamba) |
| Sequence | String: LKCYQHGKVV TCHRDMKFCY HNTGMPFRNL KLILQGCSSS CSETENNKCC STDRCNK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 5.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 103000 |
|---|---|
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: RANDOM ASSIGNMENT |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















