+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the apomorphine bound ADGRG6-Gs complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / ADGRG6 / MEMBRANE PROTEIN/IMMUNE SYSTEM / MEMBRANE PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationSchwann cell differentiation / regulation of sprouting angiogenesis / EGR2 and SOX10-mediated initiation of Schwann cell myelination / establishment of blood-brain barrier / myelination in peripheral nervous system / regulation of bone mineralization / heart trabecula formation / extracellular matrix binding / cartilage development / collagen binding ...Schwann cell differentiation / regulation of sprouting angiogenesis / EGR2 and SOX10-mediated initiation of Schwann cell myelination / establishment of blood-brain barrier / myelination in peripheral nervous system / regulation of bone mineralization / heart trabecula formation / extracellular matrix binding / cartilage development / collagen binding / laminin binding / myelination / placenta development / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / nervous system development / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / endopeptidase activity / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Ras protein signal transduction / cell surface receptor signaling pathway / Extra-nuclear estrogen signaling / cell population proliferation / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / synapse / endoplasmic reticulum membrane / protein-containing complex binding / cell surface / signal transduction / extracellular exosome / membrane / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Han S / Wu B / Zhao Q | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2026 Journal: Cell Discov / Year: 2026Title: Extra-helical allosteric binding site of apomorphine in ADGRG6 Authors: Qiu N / Xu W / Xu T / Xie W / Jiang Y / Zhong Z / Ma L / Zhao Q / Wu B / Han S | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_67602.map.gz emd_67602.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-67602-v30.xml emd-67602-v30.xml emd-67602.xml emd-67602.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_67602.png emd_67602.png | 118.8 KB | ||
| Filedesc metadata |  emd-67602.cif.gz emd-67602.cif.gz | 6.7 KB | ||
| Others |  emd_67602_half_map_1.map.gz emd_67602_half_map_1.map.gz emd_67602_half_map_2.map.gz emd_67602_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-67602 http://ftp.pdbj.org/pub/emdb/structures/EMD-67602 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-67602 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-67602 | HTTPS FTP |
-Related structure data
| Related structure data |  21duMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_67602.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_67602.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.071 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_67602_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_67602_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Apomorphine bound ADGRG6-Gs complex
| Entire | Name: Apomorphine bound ADGRG6-Gs complex |
|---|---|
| Components |
|
-Supramolecule #1: Apomorphine bound ADGRG6-Gs complex
| Supramolecule | Name: Apomorphine bound ADGRG6-Gs complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #5, #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(s) subunit alpha
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.430295 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMIE KQLQKDKQVY RATHRLLLLG ADNSGKSTIV KQMRILHGGS GGSGGTSGIF ETKFQVDKVN FHMFDVGGQ RDERRKWIQC FNDVTAIIFV VDSSDYNRLQ EALNDFKSIW NNRWLRTISV ILFLNKQDLL AEKVLAGKSK I EDYFPEFA ...String: MGCTLSAEDK AAVERSKMIE KQLQKDKQVY RATHRLLLLG ADNSGKSTIV KQMRILHGGS GGSGGTSGIF ETKFQVDKVN FHMFDVGGQ RDERRKWIQC FNDVTAIIFV VDSSDYNRLQ EALNDFKSIW NNRWLRTISV ILFLNKQDLL AEKVLAGKSK I EDYFPEFA RYTTPEDATP EPGEDPRVTR AKYFIRDEFL RISTASGDGR HYCYPHFTCA VDTENARRIF NDCRDIIQRM HL RQYELL |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.744371 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI ...String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI VTSSGDTTCA LWDIETGQQT TTFTGHTGDV MSLSLAPDTR LFVSGACDAS AKLWDVREGM CRQTFTGHES DI NAICFFP NGNAFATGSD DATCRLFDLR ADQELMTYSH DNIICGITSV SFSKSGRLLL AGYDDFNCNV WDALKADRAG VLA GHDNRV SCLGVTDDGM AVATGSWDSF LKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: nb35
| Macromolecule | Name: nb35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.540746 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGMKYLLPTA AAGLLLLAAQ PAMAQVQLQE SGGGLVQPGG SLRLSCAASG FTFSNYKMNW VRQAPGKGLE WVSDISQSGA SISYTGSVK GRFTISRDNA KNTLYLQMNS LKPEDTAVYY CARCPAPFTR DCFDVTSTTY AYRGQGTQVT VSSHHHHHHE P EA |
-Macromolecule #5: Adhesion G-protein coupled receptor G6
| Macromolecule | Name: Adhesion G-protein coupled receptor G6 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 67.947883 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNCLKEANEV ANQILNLTAD GQNLTSANIT NIVEQVKRIV NKEENIDITL GSTLMNIFSN ILSSSDSDLL ESSSEALKTI DELAFKIDL NSTSHVNITT RNLALSVSSL LPGTNAISNF SIGLPSNNES YFQMDFESGQ VDPLASVILP PNLLENLSPE D SVLVRRAQ ...String: SNCLKEANEV ANQILNLTAD GQNLTSANIT NIVEQVKRIV NKEENIDITL GSTLMNIFSN ILSSSDSDLL ESSSEALKTI DELAFKIDL NSTSHVNITT RNLALSVSSL LPGTNAISNF SIGLPSNNES YFQMDFESGQ VDPLASVILP PNLLENLSPE D SVLVRRAQ FTFFNKTGLF QDVGPQRKTL VSYVMACSIG NITIQNLKDP VQIKIKHTRT QEVHHPICAF WDLNKNKSFG GW NTSGCVA HRDSDASETV CLCNHFTHFG VLMDLPRSAS QLDARNTKVL TFISYIGCGI SAIFSAATLL TYVAFEKLRR DYP SKILMN LSTALLFLNL LFLLDGWITS FNVDGLCIAV AVLLHFFLLA TFTWMGLEAI HMYIALVKVF NTYIRRYILK FCII GWGLP ALVVSVVLAS RNNNEVYGKE SYGKEKGDEF CWIQDPVIFY VTCAGYFGVM FFLNIAMFIV VMVQICGRNG KRSNR TLRE EVLRNLRSVV SLTFLLGMTW GFAFFAWGPL NIPFMYLFSI FNSLQGLFIF IFHCAMKENV QKQWRQHLCC GRFRLE FLE VLFQGPWSHP QFEKGGGSGG GSGGSAWSHP QFEKDYKDDD DK UniProtKB: Adhesion G-protein coupled receptor G6 |
-Macromolecule #6: (6aR)-6-methyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinoline-10,1...
| Macromolecule | Name: (6aR)-6-methyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinoline-10,11-diol type: ligand / ID: 6 / Number of copies: 1 / Formula: OR9 |
|---|---|
| Molecular weight | Theoretical: 267.322 Da |
| Chemical component information | 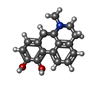 ChemComp-OR9: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































