[English] 日本語
 Yorodumi
Yorodumi- EMDB-6756: Electrostatic interaction between polyglutamylated tubulin and th... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6756 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electrostatic interaction between polyglutamylated tubulin and the nexin-dynein regulatory complex regulates flagellar motility | |||||||||
 Map data Map data | DMT structure of wild type axoneme | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
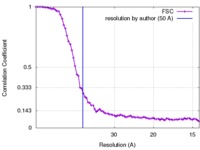
| Method | subtomogram averaging / cryo EM / Resolution: 50.0 Å | |||||||||
 Authors Authors | Kubo T / Oda T | |||||||||
 Citation Citation |  Journal: Mol Biol Cell / Year: 2017 Journal: Mol Biol Cell / Year: 2017Title: Electrostatic interaction between polyglutamylated tubulin and the nexin-dynein regulatory complex regulates flagellar motility. Authors: Tomohiro Kubo / Toshiyuki Oda /  Abstract: Tubulins undergo various posttranslational modifications. Among them, polyglutamylation is involved in the motility of eukaryotic flagella and the stability of the axonemal microtubules. However, it ...Tubulins undergo various posttranslational modifications. Among them, polyglutamylation is involved in the motility of eukaryotic flagella and the stability of the axonemal microtubules. However, it remains unclear where polyglutamylated tubulin localizes precisely within the axoneme and how tubulin polyglutamylation affects flagellar motility. In this study, we identified the three-dimensional localization of the polyglutamylated tubulin in flagella using antibody labeling and cryo-electron tomography. Polyglutamylated tubulins specifically located in close proximity to a microtubule-cross-bridging structure called the nexin-dynein regulatory complex (N-DRC). Because N-DRC is positively charged, we hypothesized that there is an electrostatic interaction between the polyglutamylated tubulin and the N-DRC, and therefore we mutated the amino acid sequences of DRC4 to modify the charge of the N-DRC. We found that both augmentation and reduction of the positive charge on DRC4 resulted in reduced flagellar motility Moreover, reduced motility in a mutant with a structurally defective N-DRC was partially restored by increasing the positive charge on DRC4. These results clearly indicate that beating motion of flagella is maintained by the electrostatic cross-bridge formed between the negatively charged polyglutamylated tubulins and the positively charged N-DRC. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6756.map.gz emd_6756.map.gz | 9.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6756-v30.xml emd-6756-v30.xml emd-6756.xml emd-6756.xml | 9.7 KB 9.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6756_fsc.xml emd_6756_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_6756.png emd_6756.png | 125.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6756 http://ftp.pdbj.org/pub/emdb/structures/EMD-6756 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6756 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6756 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6756.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6756.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DMT structure of wild type axoneme | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 7.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Outer doublet microtubule of pf2fp3 axoneme labeled with polyE2 F...
| Entire | Name: Outer doublet microtubule of pf2fp3 axoneme labeled with polyE2 Fab fragments |
|---|---|
| Components |
|
-Supramolecule #1: Outer doublet microtubule of pf2fp3 axoneme labeled with polyE2 F...
| Supramolecule | Name: Outer doublet microtubule of pf2fp3 axoneme labeled with polyE2 Fab fragments type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL |
|---|---|
| Buffer | pH: 7.2 Details: 30 mM Hepes-NaOH pH 7.2, 5 mM MgCl2, 1 mM dithiothreitol, 1 mM EGTA, 50 mM NaCl |
| Grid | Model: Homemade / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 5.5 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3100FEF |
|---|---|
| Specialist optics | Energy filter - Name: Omega / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number grids imaged: 1 / Average exposure time: 1.0 sec. / Average electron dose: 1.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: GATAN 914 HIGH TILT LIQUID NITROGEN CRYO TRANSFER TOMOGRAPHY HOLDER Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)