+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Arabidopsis thaliana URE transporter DUR3 - URE bound | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | URE transporter / homotetramer / PLANT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationurea binding / urea transmembrane transport / urea transmembrane transporter activity / symporter activity / cellular response to nitrogen starvation / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.19 Å | |||||||||
 Authors Authors | Wang YL / Lin HJ / Zhang JR / Fan MR | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Arabidopsis thaliana URE transporter DUR3 - URE bound Authors: Wang YL / Lin HJ / Zhang JR / Fan MR | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_66597.map.gz emd_66597.map.gz | 88.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-66597-v30.xml emd-66597-v30.xml emd-66597.xml emd-66597.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_66597_fsc.xml emd_66597_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_66597.png emd_66597.png | 35.1 KB | ||
| Masks |  emd_66597_msk_1.map emd_66597_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-66597.cif.gz emd-66597.cif.gz | 5.8 KB | ||
| Others |  emd_66597_half_map_1.map.gz emd_66597_half_map_1.map.gz emd_66597_half_map_2.map.gz emd_66597_half_map_2.map.gz | 164.7 MB 164.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-66597 http://ftp.pdbj.org/pub/emdb/structures/EMD-66597 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-66597 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-66597 | HTTPS FTP |
-Related structure data
| Related structure data |  9x5uMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_66597.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_66597.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.96 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_66597_msk_1.map emd_66597_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_66597_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_66597_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Arabidopsis thaliana URE transporter DUR3 - URE bound
| Entire | Name: Arabidopsis thaliana URE transporter DUR3 - URE bound |
|---|---|
| Components |
|
-Supramolecule #1: Arabidopsis thaliana URE transporter DUR3 - URE bound
| Supramolecule | Name: Arabidopsis thaliana URE transporter DUR3 - URE bound / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Urea-proton symporter DUR3
| Macromolecule | Name: Urea-proton symporter DUR3 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 80.662406 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MATCPPFDFS TKYYDGDGGC QRQSSFFGGT TVLDQGVGYA VILGFGAFFA VFTSFLVWLE KRYVGARHTS EWFNTAGRNV KTGLIASVI VSQWTWAATI LQSSNVAWQY GVSGPFWYAS GATIQVLLFG VMAIEIKRKA PNAHTVCEIV KARWGTATHI V FLVFCLAT ...String: MATCPPFDFS TKYYDGDGGC QRQSSFFGGT TVLDQGVGYA VILGFGAFFA VFTSFLVWLE KRYVGARHTS EWFNTAGRNV KTGLIASVI VSQWTWAATI LQSSNVAWQY GVSGPFWYAS GATIQVLLFG VMAIEIKRKA PNAHTVCEIV KARWGTATHI V FLVFCLAT NVVVTAMLLL GGSAVVNALT GVNLYAASFL IPLGVVVYTL AGGLKATFLA SYVHSVIVHV ALVVFVFLVY TS SKELGSP SVVYDRLKDM VAKSRSCTEP LSHHGQACGP VDGNFRGSYL TMLSSGGAVF GLINIVGNFG TVFVDNGYWV SAI AARPSS THKGYLLGGL VWFAVPFSLA TSLGLGALAL DLPISKDEAD RGLVPPATAI ALMGKSGSLL LLTMLFMAVT SAGS SELIA VSSLFTYDIY RTYINPRATG RQILKISRCA VLGFGCFMGI LAVVLNKAGV SLGWMYLAMG VLIGSAVIPI AFMLL WSKA NAFGAILGAT SGCVFGIITW LTTAKTQYGR VDLDSTGKNG PMLAGNLVAI LTGGLIHAVC SLVRPQNYDW STTREI KVV EAYASGDEDV DVPAEELREE KLRRAKAWIV KWGLVFTILI VVIWPVLSLP ARVFSRGYFW FWAIVAIAWG TIGSIVI IG LPLVESWDTI KSVCMGMFTN DRVMKKLDDL NHRLRALTMA VPEAEKIYLL ELEKTKKNDE EGVDELTSRG RDYKDDDD K WSHPQFEKGG GGSGGSAWSH PQFEK UniProtKB: Urea-proton symporter DUR3 |
-Macromolecule #2: UREA
| Macromolecule | Name: UREA / type: ligand / ID: 2 / Number of copies: 4 / Formula: URE |
|---|---|
| Molecular weight | Theoretical: 60.055 Da |
| Chemical component information | 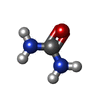 ChemComp-URE: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































