+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Zea mays URE transporter DUR3 - URE bound | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | URE transporter / homomeric tetramer / CHOLINE-BINDING PROTEIN | |||||||||
| Function / homology | Urea active transporter / urea transmembrane transport / urea transmembrane transporter activity / Sodium/solute symporter / Sodium/glucose symporter superfamily / Sodium:solute symporter family / Sodium:solute symporter family profile. / plasma membrane / Urea-proton symporter DUR3 Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.41 Å | |||||||||
 Authors Authors | Wang YL / Lin HJ / Zhang JR / Fan MR | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Zea mays URE transporter DUR3 - URE bound Authors: Wang YL / Lin HJ / Zhang JR / Fan MR | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_65093.map.gz emd_65093.map.gz | 156.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-65093-v30.xml emd-65093-v30.xml emd-65093.xml emd-65093.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_65093_fsc.xml emd_65093_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_65093.png emd_65093.png | 193.8 KB | ||
| Masks |  emd_65093_msk_1.map emd_65093_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-65093.cif.gz emd-65093.cif.gz | 5.8 KB | ||
| Others |  emd_65093_half_map_1.map.gz emd_65093_half_map_1.map.gz emd_65093_half_map_2.map.gz emd_65093_half_map_2.map.gz | 165 MB 165 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-65093 http://ftp.pdbj.org/pub/emdb/structures/EMD-65093 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-65093 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-65093 | HTTPS FTP |
-Related structure data
| Related structure data |  9vimMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_65093.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_65093.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.96 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_65093_msk_1.map emd_65093_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_65093_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_65093_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Zea mays URE transporter DUR3
| Entire | Name: Zea mays URE transporter DUR3 |
|---|---|
| Components |
|
-Supramolecule #1: Zea mays URE transporter DUR3
| Supramolecule | Name: Zea mays URE transporter DUR3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Urea-proton symporter DUR3
| Macromolecule | Name: Urea-proton symporter DUR3 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 82.153883 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAAGGAGACP PPGLGFGGEY YSVVDGACSR DGSFFGGKPV LAQAVGYAVV LGFGAFFALF TSFLVWLEKR YVGSQHTSEW FNTAGRSVK TGLIASVIVS QWTWAATILQ SSNVAWQYGV SGPFWYASGA TIQVLLFGVM AIEIKRKAPK AHTVCEIVRA R WGRAAHAV ...String: MAAGGAGACP PPGLGFGGEY YSVVDGACSR DGSFFGGKPV LAQAVGYAVV LGFGAFFALF TSFLVWLEKR YVGSQHTSEW FNTAGRSVK TGLIASVIVS QWTWAATILQ SSNVAWQYGV SGPFWYASGA TIQVLLFGVM AIEIKRKAPK AHTVCEIVRA R WGRAAHAV FLAFCLATNV IVTAMLLLGG SAVVNALTAI NVYAASFLIP LGVVVYTLAG GLKATFLASY IHSVVVHVVL VV FVFLVYT SGNRLGSPRV VHDHLTAVAS AVRNCSAPLS HPDQACGPVH GNFKGSYLTM LSSGGLVFGI INIVGNFGTV FVD NGYWMS AIAARPSSTH KGYLLGGLVW FAVPFSLATS LGLGALALDL PITAAEAAKG LVPAATATAL MGKSGSVLLL IMLF MAVTS AGSAELVAVS SLCTYDIYRT YINPDATGEQ ILRVSRATVL VFGCLMGVLA VILNLVGVSL GWLYLAMGVI IGSAV IPIA LLLLWRKANA LGAILGAVSG CVLGVTVWLS VAKVQYGRVD LDSTGRNAPM LAGNLVSILV GGAVHAACSL AWPQHY DWE SSRQITTVES VPADSDLAEE LKEERLVHAK RWIIKWGVAF TAVIAVVWPV LSLPAGRYSA GYFTMWAVIA IAWGTVG SA VIIFLPLVES WDTICKVCEG MFTNNAVYER LDEMNLRLKA IMGDMPEAEE RYQEMQKEKD LGTMEMVHPA SGTRPSIV A NDDEDNLSLA VDELTSRGRD YKDDDDKWSH PQFEKGGGGS GGSAWSHPQF EK UniProtKB: Urea-proton symporter DUR3 |
-Macromolecule #2: UREA
| Macromolecule | Name: UREA / type: ligand / ID: 2 / Number of copies: 1 / Formula: URE |
|---|---|
| Molecular weight | Theoretical: 60.055 Da |
| Chemical component information | 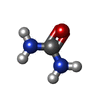 ChemComp-URE: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 130000 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































