[English] 日本語
 Yorodumi
Yorodumi- EMDB-6462: Subvolumes of individual DNA minicircles showing various shapes o... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6462 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subvolumes of individual DNA minicircles showing various shapes of the minicircles | |||||||||
 Map data Map data | Subtomogram of a DNA minicircle | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA minicircle / cryo-ET / subtomogram / supercoiled DNA / DNA minicircle shapes | |||||||||
| Biological species | unidentified (others) | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Irobalieva RN / Fogg JM / Catanese DJ / Sutthibutpong T / Chen M / Barker AK / Ludtke SJ / Harris SA / Schmid MF / Chiu W / Zechiedrich L | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2015 Journal: Nat Commun / Year: 2015Title: Structural diversity of supercoiled DNA. Authors: Rossitza N Irobalieva / Jonathan M Fogg / Daniel J Catanese / Thana Sutthibutpong / Muyuan Chen / Anna K Barker / Steven J Ludtke / Sarah A Harris / Michael F Schmid / Wah Chiu / Lynn Zechiedrich /   Abstract: By regulating access to the genetic code, DNA supercoiling strongly affects DNA metabolism. Despite its importance, however, much about supercoiled DNA (positively supercoiled DNA, in particular) ...By regulating access to the genetic code, DNA supercoiling strongly affects DNA metabolism. Despite its importance, however, much about supercoiled DNA (positively supercoiled DNA, in particular) remains unknown. Here we use electron cryo-tomography together with biochemical analyses to investigate structures of individual purified DNA minicircle topoisomers with defined degrees of supercoiling. Our results reveal that each topoisomer, negative or positive, adopts a unique and surprisingly wide distribution of three-dimensional conformations. Moreover, we uncover striking differences in how the topoisomers handle torsional stress. As negative supercoiling increases, bases are increasingly exposed. Beyond a sharp supercoiling threshold, we also detect exposed bases in positively supercoiled DNA. Molecular dynamics simulations independently confirm the conformational heterogeneity and provide atomistic insight into the flexibility of supercoiled DNA. Our integrated approach reveals the three-dimensional structures of DNA that are essential for its function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6462.map.gz emd_6462.map.gz | 14.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6462-v30.xml emd-6462-v30.xml emd-6462.xml emd-6462.xml | 9.5 KB 9.5 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6462.gif 400_6462.gif 80_6462.gif 80_6462.gif | 24.5 KB 2.4 KB | ||
| Others |  emd_6462_additional_1.map.gz emd_6462_additional_1.map.gz emd_6462_additional_2.map.gz emd_6462_additional_2.map.gz emd_6462_additional_3.map.gz emd_6462_additional_3.map.gz emd_6462_additional_4.map.gz emd_6462_additional_4.map.gz | 6.1 MB 14.4 MB 6.1 MB 6.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6462 http://ftp.pdbj.org/pub/emdb/structures/EMD-6462 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6462 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6462 | HTTPS FTP |
-Validation report
| Summary document |  emd_6462_validation.pdf.gz emd_6462_validation.pdf.gz | 78.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6462_full_validation.pdf.gz emd_6462_full_validation.pdf.gz | 77.3 KB | Display | |
| Data in XML |  emd_6462_validation.xml.gz emd_6462_validation.xml.gz | 498 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6462 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6462 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6462 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6462 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6462.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6462.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram of a DNA minicircle | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.52 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Supplemental map: emd 6462 additional 1.map
| File | emd_6462_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
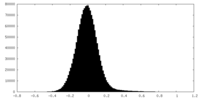
| Density Histograms |
-Supplemental map: emd 6462 additional 2.map
| File | emd_6462_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
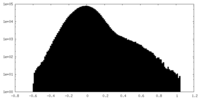
| Density Histograms |
-Supplemental map: emd 6462 additional 3.map
| File | emd_6462_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
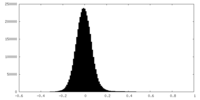
| Density Histograms |
-Supplemental map: emd 6462 additional 4.map
| File | emd_6462_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
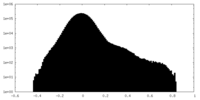
| Density Histograms |
- Sample components
Sample components
-Entire : Individual DNA minicircles of various shapes
| Entire | Name: Individual DNA minicircles of various shapes |
|---|---|
| Components |
|
-Supramolecule #1000: Individual DNA minicircles of various shapes
| Supramolecule | Name: Individual DNA minicircles of various shapes / type: sample / ID: 1000 / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 220 KDa / Method: average MW of DNA bases |
-Macromolecule #1: Minicircle DNA
| Macromolecule | Name: Minicircle DNA / type: dna / ID: 1 / Classification: DNA / Structure: DOUBLE HELIX / Synthetic?: No |
|---|---|
| Source (natural) | Organism: unidentified (others) |
| Molecular weight | Theoretical: 220 KDa |
| Sequence | String: TTTATACTAA CTTGAGCGAA ACGGGAAGGG TTTTCACCGA TATCACCGAA ACGCGCGAGG CAGCTGTATG GCATGAAAGA GTTCTTCCCG GAAAACGCGG TGGAATATTT CGTTTCCTAC TACGACTACT ATCAGCCGGA AGCCTATGTA CCGAGTTCCG ACACTTTCAT ...String: TTTATACTAA CTTGAGCGAA ACGGGAAGGG TTTTCACCGA TATCACCGAA ACGCGCGAGG CAGCTGTATG GCATGAAAGA GTTCTTCCCG GAAAACGCGG TGGAATATTT CGTTTCCTAC TACGACTACT ATCAGCCGGA AGCCTATGTA CCGAGTTCCG ACACTTTCAT TGAGAAAGAT GCCTCAGCTC TGTTACAGGT CACTAATACC ATCTAAGTAG TTGATTCATA GTGACTGCAT ATGTTGTGTT TTACAGTATT ATGTAGTCTG TTTTTTATGC AAAATCTAAT TTAATATATT GATATTTATA TCATTTTACG TTTCTCGTTC AGCTTT |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 8.3 / Details: 40 mM Tris-acetate, 10 mM CaCl2 |
| Grid | Details: 200 mesh holey carbon grids, glow-discharged for 25 seconds |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 95 K / Instrument: FEI VITROBOT MARK IV / Method: 1 blot, 2 seconds |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2200FS |
|---|---|
| Specialist optics | Energy filter - Name: In-column omega energy filter / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 25.0 eV |
| Date | Apr 19, 2011 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Average electron dose: 100 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 7.0 µm / Nominal defocus min: 5.0 µm / Nominal magnification: 25000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN / Tilt series - Axis1 - Min angle: -60 ° / Tilt series - Axis1 - Max angle: 60 ° / Tilt series - Axis1 - Angle increment: 3 ° |
- Image processing
Image processing
| Final reconstruction | Number images used: 41 |
|---|
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)