[English] 日本語
 Yorodumi
Yorodumi- EMDB-64505: Cryo-EM structure of the maize CER6-GL2 complex (inactive C222A m... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the maize CER6-GL2 complex (inactive C222A mutant) bound with malonyl-CoA | |||||||||
 Map data Map data | main map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / inactive C222A mutant / Malonyl-CoA / PLANT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationfatty acid elongase activity / acyltransferase activity, transferring groups other than amino-acyl groups / Transferases; Acyltransferases; Transferring groups other than aminoacyl groups / fatty acid biosynthetic process / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.96 Å | |||||||||
 Authors Authors | Liu Y / Zhang P | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Molecular basis of very-long-chain fatty acid elongation by the CER6-GL2 enzyme complex in plant wax biosynthesis Authors: Liu Y / Zhang P | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_64505.map.gz emd_64505.map.gz | 168.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-64505-v30.xml emd-64505-v30.xml emd-64505.xml emd-64505.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_64505_fsc.xml emd_64505_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_64505.png emd_64505.png | 112.8 KB | ||
| Filedesc metadata |  emd-64505.cif.gz emd-64505.cif.gz | 6.1 KB | ||
| Others |  emd_64505_half_map_1.map.gz emd_64505_half_map_1.map.gz emd_64505_half_map_2.map.gz emd_64505_half_map_2.map.gz | 165.1 MB 165.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-64505 http://ftp.pdbj.org/pub/emdb/structures/EMD-64505 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-64505 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-64505 | HTTPS FTP |
-Related structure data
| Related structure data |  9uu4MC  9uu3C  9uu5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_64505.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_64505.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.932 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half B
| File | emd_64505_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half A
| File | emd_64505_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of the maize CER6-GL2 complex (inactive C222A m...
| Entire | Name: Cryo-EM structure of the maize CER6-GL2 complex (inactive C222A mutant) bound with malonyl-CoA |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of the maize CER6-GL2 complex (inactive C222A m...
| Supramolecule | Name: Cryo-EM structure of the maize CER6-GL2 complex (inactive C222A mutant) bound with malonyl-CoA type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Protein ECERIFERUM 26-like
| Macromolecule | Name: Protein ECERIFERUM 26-like / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 46.86498 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDKMV FEQHEEEAVA PGAVHGHRLS TVVPSSVTGE VDYALADADL AFKLHYLRGV YYYRSGDGLA TKVLKDPMFP WLDDHFPVA GRVRRAEAEG DGAPRRPYIK CNDCGVRIVE ARCDRDMAEW IRDAAPGRIR QLCYDKVLGP ELFFSPLLYV Q ITNFKCGG ...String: DYKDDDDKMV FEQHEEEAVA PGAVHGHRLS TVVPSSVTGE VDYALADADL AFKLHYLRGV YYYRSGDGLA TKVLKDPMFP WLDDHFPVA GRVRRAEAEG DGAPRRPYIK CNDCGVRIVE ARCDRDMAEW IRDAAPGRIR QLCYDKVLGP ELFFSPLLYV Q ITNFKCGG LALGFSWAHL IGDIPSAATC FNKWAQILSG KKPEATVLTP PNQPLQGQSP AAPRSVKQVG PMEDLWLVPA GR DMACYSF HVSDAVLKKL HQQQNGRQDA AAGTFELVSA LVWQAVAKIR GDVDTVTVVR ADAAARSGKS LANEMKVGYV ESA GSSPAK TDVAELAALL AKNVVDETAA VAAFQGDVLV YGGANLTLVD MEQVDLYGLE IKGQRPVYVE YGMDGVGDEG AVLV QPDAD GRGRLVTVVL PGDEIDSLRA ALGSALHVA UniProtKB: Protein ECERIFERUM 26-like |
-Macromolecule #2: 3-ketoacyl-CoA synthase
| Macromolecule | Name: 3-ketoacyl-CoA synthase / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO EC number: Transferases; Acyltransferases; Transferring groups other than aminoacyl groups |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 56.381375 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHMPSG GVFSGSVNLK HVKLGYQYLV NHFLTLLLVP VMAATALELA RLGPGELLSL WRSLELDLVH ILCSAFLVVF VGTVYVMSR PRPVYLVDYA CYKPPASCRV PFATFMEHTR LISDDDKSVR FQTRILERSG LGEDTCLPPA NHYIPPNPSM E ASRAEAQL ...String: HHHHHHMPSG GVFSGSVNLK HVKLGYQYLV NHFLTLLLVP VMAATALELA RLGPGELLSL WRSLELDLVH ILCSAFLVVF VGTVYVMSR PRPVYLVDYA CYKPPASCRV PFATFMEHTR LISDDDKSVR FQTRILERSG LGEDTCLPPA NHYIPPNPSM E ASRAEAQL VIFSAIDDLV RRTGLKPKDI DILVVNCSLF SPTPSLSAMI INKYKLRSNI RSFNLSGMGA SAGLISIDLA RD MLQVHPN SNALVVSTEI ITPNFYQGSR RDMLLPNCLF RMGAAAILLS NRRREARRAK YRLVHVVRTH KGADDRAYRC VYE EEDEQG FSGISLSKEL MAIAGDALKS NITTIGPLVL PMSEQLLFFF RLVGRKLVNK GWRPYIPDFK LAFEHFCIHA GGRA VIDEL QKNLQLSPRH VEASRMTLHR FGNTSSSSLW YELAYIEAKG RMRRGDRVWQ IGFGSGFKCN SAVWKCLRSI KTPTN GPWD DCIHRYPVDV PEVVKL UniProtKB: 3-ketoacyl-CoA synthase |
-Macromolecule #3: MALONYL-COENZYME A
| Macromolecule | Name: MALONYL-COENZYME A / type: ligand / ID: 3 / Number of copies: 2 / Formula: MLC |
|---|---|
| Molecular weight | Theoretical: 853.58 Da |
| Chemical component information | 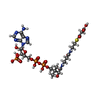 ChemComp-MLC: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































