+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | BCCP-BC Conformation of ATP-bound hPCC | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | carboxylase / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationshort-chain fatty acid catabolic process / branched-chain amino acid metabolic process / Propionyl-CoA catabolism / propionyl-CoA carboxylase / Defective HLCS causes multiple carboxylase deficiency / propionyl-CoA carboxylase activity / Biotin transport and metabolism / biotin binding / catalytic complex / Mitochondrial protein degradation ...short-chain fatty acid catabolic process / branched-chain amino acid metabolic process / Propionyl-CoA catabolism / propionyl-CoA carboxylase / Defective HLCS causes multiple carboxylase deficiency / propionyl-CoA carboxylase activity / Biotin transport and metabolism / biotin binding / catalytic complex / Mitochondrial protein degradation / fatty acid metabolic process / mitochondrial matrix / enzyme binding / mitochondrion / ATP binding / metal ion binding / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.48 Å | |||||||||
 Authors Authors | Ni FY / Yan HF / Wang QH / Ma JP | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2026 Journal: Structure / Year: 2026Title: Nanoscale conformational dynamics of human propionyl-CoA carboxylase. Authors: Huifang Yan / Fengyun Ni / Qinghua Wang / Jianpeng Ma /  Abstract: Propionyl-CoA carboxylase (PCC) is a biotin-dependent mitochondrial enzyme responsible for propionyl-CoA catabolism. Deficiencies in human PCC (hPCC) cause propionic acidemia, a severe metabolic ...Propionyl-CoA carboxylase (PCC) is a biotin-dependent mitochondrial enzyme responsible for propionyl-CoA catabolism. Deficiencies in human PCC (hPCC) cause propionic acidemia, a severe metabolic disorder driven by toxic metabolite accumulation. Despite its therapeutic relevance, the structural basis of hPCC's catalytic function remains unresolved. Here, we present high-resolution cryo-EM structures of hPCC in four distinct states, unliganded, ADP-, AMPPNP-, and ATP-bound/substrate-bound, capturing the full trajectory of the biotin carboxyl carrier protein (BCCP) domain as it translocates between active sites. Our results reinforce the crucial role of nucleotide-gated B-lid subdomain in synchronizing catalysis through coupling with BCCP movement. Structural and biochemical analysis of 10 disease-associated variants reveals how mutations disrupt key domain interfaces and dynamic motions required for activity. These new insights define the mechanistic principles governing hPCC functions, establish a structural framework for understanding PCC-related disorders, and lay the groundwork for future efforts to engineer functional replacements or modulators for metabolic therapy. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_64173.map.gz emd_64173.map.gz | 5.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-64173-v30.xml emd-64173-v30.xml emd-64173.xml emd-64173.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_64173_fsc.xml emd_64173_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_64173.png emd_64173.png | 147 KB | ||
| Masks |  emd_64173_msk_1.map emd_64173_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-64173.cif.gz emd-64173.cif.gz | 6.6 KB | ||
| Others |  emd_64173_half_map_1.map.gz emd_64173_half_map_1.map.gz emd_64173_half_map_2.map.gz emd_64173_half_map_2.map.gz | 200.2 MB 200.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-64173 http://ftp.pdbj.org/pub/emdb/structures/EMD-64173 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-64173 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-64173 | HTTPS FTP |
-Validation report
| Summary document |  emd_64173_validation.pdf.gz emd_64173_validation.pdf.gz | 924.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_64173_full_validation.pdf.gz emd_64173_full_validation.pdf.gz | 923.8 KB | Display | |
| Data in XML |  emd_64173_validation.xml.gz emd_64173_validation.xml.gz | 21.7 KB | Display | |
| Data in CIF |  emd_64173_validation.cif.gz emd_64173_validation.cif.gz | 28.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-64173 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-64173 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-64173 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-64173 | HTTPS FTP |
-Related structure data
| Related structure data |  9uhrMC  8zuxC  8zuyC  8zuzC  8zv0C  8zv1C  8zv2C  8zv3C  8zv4C  8zv5C  8zv6C  9uh1C  9uh2C  9uh5C  9uh6C  9uh8C  9uhbC  9uhsC  9uhyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_64173.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_64173.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_64173_msk_1.map emd_64173_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_64173_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_64173_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Propionyl-CoA carboxylase
| Entire | Name: Propionyl-CoA carboxylase |
|---|---|
| Components |
|
-Supramolecule #1: Propionyl-CoA carboxylase
| Supramolecule | Name: Propionyl-CoA carboxylase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Propionyl-CoA carboxylase alpha chain, mitochondrial
| Macromolecule | Name: Propionyl-CoA carboxylase alpha chain, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: propionyl-CoA carboxylase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 80.161922 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAGFWVGTAP LVAAGRRGRW PPQQLMLSAA LRTLKHVLYY SRQCLMVSRN LGSVGYDPNE KTFDKILVAN RGEIACRVIR TCKKMGIKT VAIHSDVDAS SVHVKMADEA VCVGPAPTSK SYLNMDAIME AIKKTRAQAV HPGYGFLSEN KEFARCLAAE D VVFIGPDT ...String: MAGFWVGTAP LVAAGRRGRW PPQQLMLSAA LRTLKHVLYY SRQCLMVSRN LGSVGYDPNE KTFDKILVAN RGEIACRVIR TCKKMGIKT VAIHSDVDAS SVHVKMADEA VCVGPAPTSK SYLNMDAIME AIKKTRAQAV HPGYGFLSEN KEFARCLAAE D VVFIGPDT HAIQAMGDKI ESKLLAKKAE VNTIPGFDGV VKDAEEAVRI AREIGYPVMI KASAGGGGKG MRIAWDDEET RD GFRLSSQ EAASSFGDDR LLIEKFIDNP RHIEIQVLGD KHGNALWLNE RECSIQRRNQ KVVEEAPSIF LDAETRRAMG EQA VALARA VKYSSAGTVE FLVDSKKNFY FLEMNTRLQV EHPVTECITG LDLVQEMIRV AKGYPLRHKQ ADIRINGWAV ECRV YAEDP YKSFGLPSIG RLSQYQEPLH LPGVRVDSGI QPGSDISIYY DPMISKLITY GSDRTEALKR MADALDNYVI RGVTH NIAL LREVIINSRF VKGDISTKFL SDVYPDGFKG HMLTKSEKNQ LLAIASSLFV AFQLRAQHFQ ENSRMPVIKP DIANWE LSV KLHDKVHTVV ASNNGSVFSV EVDGSKLNVT STWNLASPLL SVSVDGTQRT VQCLSREAGG NMSIQFLGTV YKVNILT RL AAELNKFMLE KVTEDTSSVL RSPMPGVVVA VSVKPGDAVA EGQEICVIEA MKMQNSMTAG KTGTVKSVHC QAGDTVGE G DLLVELE UniProtKB: Propionyl-CoA carboxylase alpha chain, mitochondrial |
-Macromolecule #2: Propionyl-CoA carboxylase beta chain, mitochondrial
| Macromolecule | Name: Propionyl-CoA carboxylase beta chain, mitochondrial / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: propionyl-CoA carboxylase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 27.095178 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DPSDRLVPEL DTIVPLESTK AYNMVDIIHS VVDEREFFEI MPNYAKNIIV GFARMNGRTV GIVGNQPKVA SGCLDINSSV KGARFVRFC DAFNIPLITF VDVPGFLPGT AQEYGGIIRH GAKLLYAFAE ATVPKVTVIT RKAYGGAYDV MSSKHLCGDT N YAWPTAEI ...String: DPSDRLVPEL DTIVPLESTK AYNMVDIIHS VVDEREFFEI MPNYAKNIIV GFARMNGRTV GIVGNQPKVA SGCLDINSSV KGARFVRFC DAFNIPLITF VDVPGFLPGT AQEYGGIIRH GAKLLYAFAE ATVPKVTVIT RKAYGGAYDV MSSKHLCGDT N YAWPTAEI AVMGAKGAVE IIFKGHENVE AAQAEYIEKF ANPFPAAVRG FVDDIIQPSS TRARICCDLD VLASKKVQRP WR KHANIPL UniProtKB: Propionyl-CoA carboxylase beta chain, mitochondrial |
-Macromolecule #3: Propionyl-CoA carboxylase beta chain, mitochondrial
| Macromolecule | Name: Propionyl-CoA carboxylase beta chain, mitochondrial / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: propionyl-CoA carboxylase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 31.20749 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAAALRVAAV GARLSVLASG LRAAVRSLCS QATSVNERIE NKRRTALLGG GQRRIDAQHK RGKLTARERI SLLLDPGSFV ESDMFVEHR CADFGMAADK NKFPGDSVVT GRGRINGRLV YVFSQDFTVF GGSLSGAHAQ KICKIMDQAI TVGAPVIGLN D SGGARIQE ...String: MAAALRVAAV GARLSVLASG LRAAVRSLCS QATSVNERIE NKRRTALLGG GQRRIDAQHK RGKLTARERI SLLLDPGSFV ESDMFVEHR CADFGMAADK NKFPGDSVVT GRGRINGRLV YVFSQDFTVF GGSLSGAHAQ KICKIMDQAI TVGAPVIGLN D SGGARIQE GVESLAGYAD IFLRNVTASG VIPQISLIMG PCAGGAVYSP ALTDFTFMVK DTSYLFITGP DVVKSVTNED VT QEELGGA KTHTTMSGVA HRAFENDVDA LCNLRDFFNY LPLSSQDPAP VRECH UniProtKB: Propionyl-CoA carboxylase beta chain, mitochondrial |
-Macromolecule #4: BIOTIN
| Macromolecule | Name: BIOTIN / type: ligand / ID: 4 / Number of copies: 1 / Formula: BTN |
|---|---|
| Molecular weight | Theoretical: 244.311 Da |
| Chemical component information | 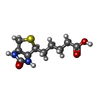 ChemComp-BTN: |
-Macromolecule #5: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #7: BICARBONATE ION
| Macromolecule | Name: BICARBONATE ION / type: ligand / ID: 7 / Number of copies: 1 / Formula: BCT |
|---|---|
| Molecular weight | Theoretical: 61.017 Da |
| Chemical component information |  ChemComp-BCT: |
-Macromolecule #8: propionyl Coenzyme A
| Macromolecule | Name: propionyl Coenzyme A / type: ligand / ID: 8 / Number of copies: 1 / Formula: 1VU |
|---|---|
| Molecular weight | Theoretical: 823.597 Da |
| Chemical component information |  ChemComp-191: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































