[English] 日本語
 Yorodumi
Yorodumi- EMDB-64089: Cryo-EM structure of the CT-PT-BCCP domain of pyruvate carboxylas... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the CT-PT-BCCP domain of pyruvate carboxylase from Mycobacterium tuberculosis | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Pyruvate Carboxylase / BCCP / CT / Mycobacterium tuberculosis / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationpyruvate carboxylase / pyruvate carboxylase activity / biotin carboxylase activity / gluconeogenesis / ATP binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Singh A / Raza M / Singh S / Das U | |||||||||
| Funding support |  India, 1 items India, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of Pyruvate carboxylase from Mycobacterium tuberculosis in complex with Acetyl-CoA and ADP Authors: Singh A / Raza M / Singh S / Das U | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_64089.map.gz emd_64089.map.gz | 89.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-64089-v30.xml emd-64089-v30.xml emd-64089.xml emd-64089.xml | 27 KB 27 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_64089_fsc.xml emd_64089_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_64089.png emd_64089.png | 123.6 KB | ||
| Masks |  emd_64089_msk_1.map emd_64089_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-64089.cif.gz emd-64089.cif.gz | 7.6 KB | ||
| Others |  emd_64089_additional_1.map.gz emd_64089_additional_1.map.gz emd_64089_half_map_1.map.gz emd_64089_half_map_1.map.gz emd_64089_half_map_2.map.gz emd_64089_half_map_2.map.gz | 149.1 MB 165.4 MB 165.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-64089 http://ftp.pdbj.org/pub/emdb/structures/EMD-64089 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-64089 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-64089 | HTTPS FTP |
-Related structure data
| Related structure data |  9ueqMC  9ubgC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_64089.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_64089.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_64089_msk_1.map emd_64089_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: DeepEMhancer sharpened map
| File | emd_64089_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half Map A
| File | emd_64089_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half Map B
| File | emd_64089_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Pyruvate carboxylase CT and BCCP domain
| Entire | Name: Pyruvate carboxylase CT and BCCP domain |
|---|---|
| Components |
|
-Supramolecule #1: Pyruvate carboxylase CT and BCCP domain
| Supramolecule | Name: Pyruvate carboxylase CT and BCCP domain / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
| Molecular weight | Theoretical: 482 KDa |
-Macromolecule #1: Pyruvate carboxylase
| Macromolecule | Name: Pyruvate carboxylase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: pyruvate carboxylase |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
| Molecular weight | Theoretical: 71.522039 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ADRGTKILNF LADVTVNNPY GSRPSTIYPD DKLPDLDLRA APPAGSKQRL VKLGPEGFAR WLRESAAVGV TDTTFRDAHQ SLLATRVRT SGLSRVAPYL ARTMPQLLSV ECWGGATYDV ALRFLKEDPW ERLATLRAAM PNICLQMLLR GRNTVGYTPY P EIVTSAFV ...String: ADRGTKILNF LADVTVNNPY GSRPSTIYPD DKLPDLDLRA APPAGSKQRL VKLGPEGFAR WLRESAAVGV TDTTFRDAHQ SLLATRVRT SGLSRVAPYL ARTMPQLLSV ECWGGATYDV ALRFLKEDPW ERLATLRAAM PNICLQMLLR GRNTVGYTPY P EIVTSAFV QEATATGIDI FRIFDALNNI ESMRPAIDAV RETGSAIAEV AMCYTGDLTD PGEQLYTLDY YLKLAEQIVD AG AHVLAIK DMAGLLRPPA AQRLVSALRS RFDLPVHLHT HDTPGGQLAS YVAAWHAGAD AVDGAAAPLA GTTSQPALSS IVA AAAHTE YDTGLSLSAV CALEPYWEAL RKVYAPFESG LPGPTGRVYH HEIPGGQLSN LRQQAIALGL GDRFEEIEEA YAGA DRVLG RLVKVTPTSK VVGDLALALV GAGVSADEFA SDPARFGIPE SVLGFLRGEL GDPPGGWPEP LRTAALAGRG AARPT AQLA ADDEIALSSV GAKRQATLNR LLFPSPTKEF NEHREAYGDT SQLSANQFFY GLRQGEEHRV KLERGVELLI GLEAIS EPD ERGMRTVMCI LNGQLRPVLV RDRSIASAVP AAEKADRGNP GHIAAPFAGV VTVGVCVGER VGAGQTIATI EAMKMEA PI TAPVAGTVER VAVSDTAQVE GGDLLVVVS UniProtKB: Pyruvate carboxylase |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #3: BIOTIN
| Macromolecule | Name: BIOTIN / type: ligand / ID: 3 / Number of copies: 2 / Formula: BTN |
|---|---|
| Molecular weight | Theoretical: 244.311 Da |
| Chemical component information | 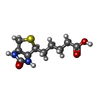 ChemComp-BTN: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 3 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Software | Name: EPU |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 6780 / Average exposure time: 1.04 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















































