[English] 日本語
 Yorodumi
Yorodumi- EMDB-64050: cryo-EM structure of M1 muscarinic acetylcholine receptor-alpha5 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of M1 muscarinic acetylcholine receptor-alpha5 helix of G11 protein complex bound to iperoxo and nanobody Nb1B4 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / active-state / nanobody / de novo protein / MEMBRANE PROTEIN/IMMUNE SYSTEM / MEMBRANE PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of melanocyte differentiation / phospholipase C-activating G protein-coupled acetylcholine receptor signaling pathway / endothelin receptor signaling pathway / Fatty Acids bound to GPR40 (FFAR1) regulate insulin secretion / Acetylcholine regulates insulin secretion / developmental pigmentation / phospholipase C-activating dopamine receptor signaling pathway / cellular response to pH / PLC beta mediated events / entrainment of circadian clock ...regulation of melanocyte differentiation / phospholipase C-activating G protein-coupled acetylcholine receptor signaling pathway / endothelin receptor signaling pathway / Fatty Acids bound to GPR40 (FFAR1) regulate insulin secretion / Acetylcholine regulates insulin secretion / developmental pigmentation / phospholipase C-activating dopamine receptor signaling pathway / cellular response to pH / PLC beta mediated events / entrainment of circadian clock / cranial skeletal system development / ligand-gated ion channel signaling pathway / phototransduction, visible light / action potential / photoreceptor outer segment / enzyme regulator activity / Turbulent (oscillatory, disturbed) flow shear stress activates signaling by PIEZO1 and integrins in endothelial cells / skeletal system development / positive regulation of insulin secretion / G protein-coupled receptor binding / regulation of blood pressure / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein beta/gamma-subunit complex binding / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / ADP signalling through P2Y purinoceptor 1 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / heterotrimeric G-protein complex / Thrombin signalling through proteinase activated receptors (PARs) / heart development / G protein activity / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / lysosomal membrane / GTPase activity / synapse / GTP binding / signal transduction / extracellular exosome / metal ion binding / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.88 Å | |||||||||
 Authors Authors | Zhang X / Gao K / Liu X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: cryo-EM structure of M1 muscarinic acetylcholine receptor-alpha5 helix of G11 protein complex bound to iperoxo and nanobody Nb1B4 Authors: Zhang X / Gao K / Liu X | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_64050.map.gz emd_64050.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-64050-v30.xml emd-64050-v30.xml emd-64050.xml emd-64050.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_64050.png emd_64050.png | 55 KB | ||
| Masks |  emd_64050_msk_1.map emd_64050_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-64050.cif.gz emd-64050.cif.gz | 6.1 KB | ||
| Others |  emd_64050_half_map_1.map.gz emd_64050_half_map_1.map.gz emd_64050_half_map_2.map.gz emd_64050_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-64050 http://ftp.pdbj.org/pub/emdb/structures/EMD-64050 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-64050 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-64050 | HTTPS FTP |
-Related structure data
| Related structure data |  9ucpMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_64050.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_64050.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0825 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_64050_msk_1.map emd_64050_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_64050_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_64050_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : cryo-EM structure of M1 muscarinic acetylcholine receptor-alpha5 ...
| Entire | Name: cryo-EM structure of M1 muscarinic acetylcholine receptor-alpha5 helix of G11 protein complex bound to iperoxo and nanobody Nb1B4/de novo design fusion protein |
|---|---|
| Components |
|
-Supramolecule #1: cryo-EM structure of M1 muscarinic acetylcholine receptor-alpha5 ...
| Supramolecule | Name: cryo-EM structure of M1 muscarinic acetylcholine receptor-alpha5 helix of G11 protein complex bound to iperoxo and nanobody Nb1B4/de novo design fusion protein type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: M1 muscarinic acetylcholine receptor, de novo design protein
| Macromolecule | Name: M1 muscarinic acetylcholine receptor, de novo design protein type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 49.317816 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDAAA QTSAPPAVSP QITVLAPGKG PWQVAFIGIT TGLLSLATVT GNLLVLISFK VNTELKTVNN YFLLSLACAD LIIGTFSMN LYTTYLLMGH WALGTLACDL WLALDYVASN ASVMNLLLIS FDRYFSVTRP LSYRAKRTPR RAALMIGLAW L VSFVLWAP ...String: DYKDDDDAAA QTSAPPAVSP QITVLAPGKG PWQVAFIGIT TGLLSLATVT GNLLVLISFK VNTELKTVNN YFLLSLACAD LIIGTFSMN LYTTYLLMGH WALGTLACDL WLALDYVASN ASVMNLLLIS FDRYFSVTRP LSYRAKRTPR RAALMIGLAW L VSFVLWAP AILFWQYLVG ERTVLAGQCY IQFLSQPIIT FGTAMAAFYL PVTVMCTLYW RIYRETKRAG ERLAKLLEKF EA LPLEDIV AALKALLATN RPEIQLAVKT IVENFPEIKK EAEKLTPEQK AKLAALEAQL ADLPEELRKV LLSMYLTGLL YGG SEENRK RAIEKAARTL SAILLAFILT WTPYNIMVLV STFCKDCVPE TLWELGYWLC YVNSTINPMC YALCNKAFRD TFRL LLLCR WDKRRWRKIP KRPGSVHRTP SRQCHHHHHH |
-Macromolecule #2: Guanine nucleotide-binding protein subunit alpha-11
| Macromolecule | Name: Guanine nucleotide-binding protein subunit alpha-11 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 3.15266 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TPENIRFVFA AVKDTILQLN LKEYNLV UniProtKB: Guanine nucleotide-binding protein subunit alpha-11 |
-Macromolecule #3: Nanobody Nb1B4
| Macromolecule | Name: Nanobody Nb1B4 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.766307 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLQESGGG LVQAGGSLRL SCAASGSIFY GYYMGWYRQA PGKEREFVAT INYGASTNYA DSVKGRFTIS RDNAKNTVYL QMNSLKPED TAVYYCAVRR VYVYVQRYYH YYWGQGTQVT VSS |
-Macromolecule #4: 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium
| Macromolecule | Name: 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium type: ligand / ID: 4 / Number of copies: 1 / Formula: IXO |
|---|---|
| Molecular weight | Theoretical: 197.254 Da |
| Chemical component information | 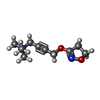 ChemComp-IXO: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.1 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)












































