+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human OAT4 in complex with DHEAS | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transporter / OAT4 / organic anion transporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationOrganic anion transport by SLC22 transporters / : / prostaglandin transport / prostaglandin transmembrane transporter activity / urate metabolic process / solute:inorganic anion antiporter activity / organic anion transport / : / monoatomic ion transport / basal plasma membrane ...Organic anion transport by SLC22 transporters / : / prostaglandin transport / prostaglandin transmembrane transporter activity / urate metabolic process / solute:inorganic anion antiporter activity / organic anion transport / : / monoatomic ion transport / basal plasma membrane / apical plasma membrane / external side of plasma membrane / extracellular exosome / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.98 Å | |||||||||
 Authors Authors | Zhang W / Chu Y / Zhang Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of human OAT4 in complex with DHEAS Authors: Zhang W / Chu Y / Zhang Y | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_63867.map.gz emd_63867.map.gz | 38.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-63867-v30.xml emd-63867-v30.xml emd-63867.xml emd-63867.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_63867_fsc.xml emd_63867_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_63867.png emd_63867.png | 50 KB | ||
| Filedesc metadata |  emd-63867.cif.gz emd-63867.cif.gz | 5.7 KB | ||
| Others |  emd_63867_half_map_1.map.gz emd_63867_half_map_1.map.gz emd_63867_half_map_2.map.gz emd_63867_half_map_2.map.gz | 37.8 MB 37.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-63867 http://ftp.pdbj.org/pub/emdb/structures/EMD-63867 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-63867 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-63867 | HTTPS FTP |
-Related structure data
| Related structure data |  9u5aMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_63867.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_63867.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.932 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_63867_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_63867_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : OAT4
| Entire | Name: OAT4 |
|---|---|
| Components |
|
-Supramolecule #1: OAT4
| Supramolecule | Name: OAT4 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 22 member 11
| Macromolecule | Name: Solute carrier family 22 member 11 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 62.769094 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAFSKLLEQA GGVGLFQTLQ VLTFILPCLM IPSQMLLENF SAAIPGHRCW THMLDNGSAV STNMTPKALL TISIPPGPNQ GPHQCRRFR QPQWQLLDPN ATATSWSEAD TEPCVDGWVY DRSVFTSTIV AKWDLVCSSQ GLKPLSQSIF MSGILVGSFI W GLLSYRFG ...String: MAFSKLLEQA GGVGLFQTLQ VLTFILPCLM IPSQMLLENF SAAIPGHRCW THMLDNGSAV STNMTPKALL TISIPPGPNQ GPHQCRRFR QPQWQLLDPN ATATSWSEAD TEPCVDGWVY DRSVFTSTIV AKWDLVCSSQ GLKPLSQSIF MSGILVGSFI W GLLSYRFG RKPMLSWCCL QLAVAGTSTI FAPTFVIYCG LRFVAAFGMA GIFLSSLTLM VEWTTTSRRA VTMTVVGCAF SA GQAALGG LAFALRDWRT LQLAASVPFF AISLISWWLP ESARWLIIKG KPDQALQELR KVARINGHKE AKNLTIEVLM SSV KEEVAS AKEPRSVLDL FCVPVLRWRS CAMLVVNFSL LISYYGLVFD LQSLGRDIFL LQALFGAVDF LGRATTALLL SFLG RRTIQ AGSQAMAGLA ILANMLVPQD LQTLRVVFAV LGKGCFGISL TCLTIYKAEL FPTPVRMTAD GILHTVGRLG AMMGP LILM SRQALPLLPP LLYGVISIAS SLVVLFFLPE TQGLPLPDTI QDLESQKSTA AQGNRQEAVT VESTSLHHHH HHHHHH GSV EDYKDDDDK UniProtKB: Solute carrier family 22 member 11 |
-Macromolecule #2: 17-oxoandrost-5-en-3beta-yl hydrogen sulfate
| Macromolecule | Name: 17-oxoandrost-5-en-3beta-yl hydrogen sulfate / type: ligand / ID: 2 / Number of copies: 1 / Formula: ZWY |
|---|---|
| Molecular weight | Theoretical: 368.488 Da |
| Chemical component information | 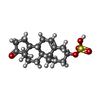 ChemComp-ZWY: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Average electron dose: 46.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.9 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































