+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Gi-bound GPCR | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / PROTEIN / Antagonist / MEMBRANE / MEMBRANE PROTEIN-IMMUNE SYSTEM complex / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationgroup III metabotropic glutamate receptor activity / adenylate cyclase-inhibiting G protein-coupled glutamate receptor signaling pathway / G protein-coupled glutamate receptor signaling pathway / Class C/3 (Metabotropic glutamate/pheromone receptors) / glutamate receptor activity / adenylate cyclase inhibitor activity / regulation of synaptic transmission, glutamatergic / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway ...group III metabotropic glutamate receptor activity / adenylate cyclase-inhibiting G protein-coupled glutamate receptor signaling pathway / G protein-coupled glutamate receptor signaling pathway / Class C/3 (Metabotropic glutamate/pheromone receptors) / glutamate receptor activity / adenylate cyclase inhibitor activity / regulation of synaptic transmission, glutamatergic / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / D2 dopamine receptor binding / response to prostaglandin E / adenylate cyclase regulator activity / G protein-coupled serotonin receptor binding / adenylate cyclase-inhibiting serotonin receptor signaling pathway / visual perception / cellular response to forskolin / regulation of mitotic spindle organization / Regulation of insulin secretion / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / G protein-coupled receptor activity / response to peptide hormone / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein beta/gamma-subunit complex binding / centriolar satellite / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through CDC42 / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / GDP binding / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / cellular response to catecholamine stimulus / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / Inactivation, recovery and regulation of the phototransduction cascade / cellular response to prostaglandin E stimulus / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / sensory perception of taste / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / G protein activity / GTPase binding / Ca2+ pathway / midbody / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / cell cortex / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / ciliary basal body / G protein-coupled receptor signaling pathway / lysosomal membrane / cell division / GTPase activity / synapse / centrosome / GTP binding / protein-containing complex binding / nucleolus / magnesium ion binding / Golgi apparatus / signal transduction / extracellular exosome / nucleoplasm / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.97 Å | |||||||||
 Authors Authors | Zhao J / Zhao C / Sun H / Shao ZH / Sun JP | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2025 Journal: Mol Cell / Year: 2025Title: Structural characterization of five functional states of metabotropic glutamate receptor 8. Authors: Jie Zhao / Yue Deng / Zheng Xu / Chanjuan Xu / Chang Zhao / Ziyan Li / Hui Sun / Xiaowen Tian / Yuxuan Song / Marta Cimadevila / Heli Wang / Yuxuan Liu / Xiaoyu Zhang / Yiyang Chen / Suyue ...Authors: Jie Zhao / Yue Deng / Zheng Xu / Chanjuan Xu / Chang Zhao / Ziyan Li / Hui Sun / Xiaowen Tian / Yuxuan Song / Marta Cimadevila / Heli Wang / Yuxuan Liu / Xiaoyu Zhang / Yiyang Chen / Suyue Sun / Xihao Yong / Lantian Su / Yixiao He / Yi Zhong / Hao Yang / Jean-Philippe Pin / Wei Yan / Zhenhua Shao / Jianfeng Liu /   Abstract: Metabotropic glutamate receptors (mGluRs) are dimeric class C G protein-coupled receptors, which play crucial roles in brain physiology and pathology. Among them, mGlu8 is the least characterized, ...Metabotropic glutamate receptors (mGluRs) are dimeric class C G protein-coupled receptors, which play crucial roles in brain physiology and pathology. Among them, mGlu8 is the least characterized, though it is physiologically important. While recognized to signal via G proteins, the involvement of β-arrestin is unknown. Here, we found that both mGlu8 agonists and positive allosteric modulators (PAMs) activate G signaling, but mainly agonists induce β-arrestin recruitment. We solved five human mGlu8 cryo-electron microscopy (cryo-EM) structures in various states: apo, antagonist-bound, agonist + PAM-bound, agonist + PAM-bound with G protein, and agonist-bound with β-arrestin1 states. They revealed a unique PAM-binding pocket at the extracellular side of the TM6/TM7 interface. Agonist and PAM promote active mGlu8 association with one G protein asymmetrically (2:1), while two β-arrestin1 can interact symmetrically (2:2) to both subunits of an inactive dimer state to promote constitutive internalization. These findings elucidate how mGlu8 selectively engages transducers, offering insights into its signaling capabilities and selective drug development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_63770.map.gz emd_63770.map.gz | 210.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-63770-v30.xml emd-63770-v30.xml emd-63770.xml emd-63770.xml | 26.8 KB 26.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_63770.png emd_63770.png | 26.5 KB | ||
| Filedesc metadata |  emd-63770.cif.gz emd-63770.cif.gz | 7.2 KB | ||
| Others |  emd_63770_additional_1.map.gz emd_63770_additional_1.map.gz emd_63770_additional_2.map.gz emd_63770_additional_2.map.gz emd_63770_additional_3.map.gz emd_63770_additional_3.map.gz emd_63770_half_map_1.map.gz emd_63770_half_map_1.map.gz emd_63770_half_map_2.map.gz emd_63770_half_map_2.map.gz | 367.5 MB 367.9 MB 367.5 MB 391.5 MB 391.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-63770 http://ftp.pdbj.org/pub/emdb/structures/EMD-63770 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-63770 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-63770 | HTTPS FTP |
-Validation report
| Summary document |  emd_63770_validation.pdf.gz emd_63770_validation.pdf.gz | 934.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_63770_full_validation.pdf.gz emd_63770_full_validation.pdf.gz | 934.2 KB | Display | |
| Data in XML |  emd_63770_validation.xml.gz emd_63770_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_63770_validation.cif.gz emd_63770_validation.cif.gz | 19.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-63770 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-63770 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-63770 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-63770 | HTTPS FTP |
-Related structure data
| Related structure data |  9mb9MC  9mbaC  9mbbC  9mbcC  9mbdC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_63770.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_63770.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.75 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Local refinement focused on mGluR8 VFT and CRD
| File | emd_63770_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local refinement focused on mGluR8 VFT and CRD | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Local refinement focused on mGluR8 TMD
| File | emd_63770_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local refinement focused on mGluR8 TMD | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Local refinement focused on Gi1
| File | emd_63770_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local refinement focused on Gi1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_63770_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_63770_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : GPCR bound to Gi1 heterotrimer
| Entire | Name: GPCR bound to Gi1 heterotrimer |
|---|---|
| Components |
|
-Supramolecule #1: GPCR bound to Gi1 heterotrimer
| Supramolecule | Name: GPCR bound to Gi1 heterotrimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Metabotropic glutamate receptor 8
| Macromolecule | Name: Metabotropic glutamate receptor 8 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 101.858414 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVCEGKRSAS CPCFFLLTAK FYWILTMMQR THSQEYAHSI RVDGDIILGG LFPVHAKGER GVPCGELKKE KGIHRLEAML YAIDQINKD PDLLSNITLG VRILDTCSRD TYALEQSLTF VQALIEKDAS DVKCANGDPP IFTKPDKISG VIGAAASSVS I MVANILRL ...String: MVCEGKRSAS CPCFFLLTAK FYWILTMMQR THSQEYAHSI RVDGDIILGG LFPVHAKGER GVPCGELKKE KGIHRLEAML YAIDQINKD PDLLSNITLG VRILDTCSRD TYALEQSLTF VQALIEKDAS DVKCANGDPP IFTKPDKISG VIGAAASSVS I MVANILRL FKIPQISYAS TAPELSDNTR YDFFSRVVPP DSYQAQAMVD IVTALGWNYV STLASEGNYG ESGVEAFTQI SR EIGGVCI AQSQKIPREP RPGEFEKIIK RLLETPNARA VIMFANEDDI RRILEAAKKL NQSGHFLWIG SDSWGSKIAP VYQ QEEIAE GAVTILPKRA SIDGFDRYFR SRTLANNRRN VWFAEFWEEN FGCKLGSHGK RNSHIKKCTG LERIARDSSY EQEG KVQFV IDAVYSMAYA LHNMHKDLCP GYIGLCPRMS TIDGKELLGY IRAVNFNGSA GTPVTFNENG DAPGRYDIFQ YQITN KSTE YKVIGHWTNQ LHLKVEDMQW AHREHTHPAS VCSLPCKPGE RKKTVKGVPC CWHCERCEGY NYQVDELSCE LCPLDQ RPN MNRTGCQLIP IIKLEWHSPW AVVPVFVAIL GIIATTFVIV TFVRYNDTPI VRASGRELSY VLLTGIFLCY SITFLMI AA PDTIICSFRR VFLGLGMCFS YAALLTKTNR IHRIFEQGKK SVTAPKFISP ASQLVITFSL ISVQLLGVFV WFVVDPPH I IIDYGEQRTL DPEKARGVLK CDISDLSLIC SLGYSILLMV TCTVYAIKTR GVPETFNEAK PIGFTMYTTC IIWLAFIPI FFGTAQSAEK MYIQTTTLTV SMSLSASVSL GMLYMPKVYI IIFHPEQNVQ KRKRSFKAVV TAATMQSKLI QKGNDRPNGE VKSELCESL ETNTSSTKTT YISYSNHSI UniProtKB: Metabotropic glutamate receptor 8 |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.414047 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHASM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.573988 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSSGSELDQL RQEAEQLKNQ IRDARKACAD ATLSQITNNI DPVGRIQMRT RRTLRGHLAK IYAMHWGTDS RLLVSASQDG KLIIWDSYT TNKVHAIPLR SSWVMTCAYA PSGNYVACGG LDNICSIYNL KTREGNVRVS RELAGHTGYL SCCRFLDDNQ I VTSSGDTT ...String: GSSGSELDQL RQEAEQLKNQ IRDARKACAD ATLSQITNNI DPVGRIQMRT RRTLRGHLAK IYAMHWGTDS RLLVSASQDG KLIIWDSYT TNKVHAIPLR SSWVMTCAYA PSGNYVACGG LDNICSIYNL KTREGNVRVS RELAGHTGYL SCCRFLDDNQ I VTSSGDTT CALWDIETGQ QTTTFTGHTG DVMSLSLAPD TRLFVSGACD ASAKLWDVRE GMCRQTFTGH ESDINAICFF PN GNAFATG SDDATCRLFD LRADQELMTY SHDNIICGIT SVSFSKSGRL LLAGYDDFNC NVWDALKADR AGVLAGHDNR VSC LGVTDD GMAVATGSWD SFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: 4-[(S)-amino(carboxy)methyl]benzene-1,2-dicarboxylic acid
| Macromolecule | Name: 4-[(S)-amino(carboxy)methyl]benzene-1,2-dicarboxylic acid type: ligand / ID: 5 / Number of copies: 2 / Formula: HVG |
|---|---|
| Molecular weight | Theoretical: 239.182 Da |
| Chemical component information | 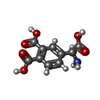 ChemComp-HVG: |
-Macromolecule #6: ~{N}-[3-chloranyl-4-(5-chloranylpyridin-2-yl)oxy-phenyl]pyridine-...
| Macromolecule | Name: ~{N}-[3-chloranyl-4-(5-chloranylpyridin-2-yl)oxy-phenyl]pyridine-2-carboxamide type: ligand / ID: 6 / Number of copies: 1 / Formula: A1ENS |
|---|---|
| Molecular weight | Theoretical: 360.194 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.3 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




























































